Store additional data
Please note that additional data storage is in a work in progress state with limited support.
Prior to version 0.3.0 the molecule stores only supported storing and retrieving a single arbitrary string for each molecule (referred as ID). With the version 0.3.2 additional properties stored when an overlap analysis is calculated can be used in overlap visualization.
Limited support
- Additional data can be attached only to molecules imported by
calculateOverlap.sh. - Additional data can be displayed on molecule details dialog of the molecules page.
- Numeric additional data can be used on overlap analysis visualization.
- REST API makes querying possible.
Specify additional data
Tool calculateOverlap.sh can add extra data to the query and target sets too. See the command line help of options -qprop <SPEC> and -tprop <SPEC>. Property specifications are applied to each processed molecule and are stored in the serialized files.
Specification format
Each specification parameter is a string in the format of <NAME>:<VALUETYPE>:<CALCTYPE>[:<CALCSPEC> where
-
NAMEIs the name which identifies the stored additional property for the query or the target molecule sets. Values must be unique across the molecule sets, so two stored property can not have the same name assigned. -
<VALUETYPE>Expected/interpreted type of the stored property. Must be explicitly specified. Applicable types:int: 32 bit signed integer valuelong: 64 bit signed integer valuedouble: 64 bit floating point valuestring: Unicode character sequence
-
<CALCTYPE>: Calculator/extractor to be used to get the value for each input molecules. <CALCSPEC>: Optional parameter/argument passed to the calculator/extractor.
Currently supported calculators, their value types and parameter strings:
<CALCTYPE> |
Value types | Description and argument (<CALSPEC>) interpretation |
|---|---|---|
molname |
string |
Use the name stored in the input molecule. No argument accepted. |
sdfprop |
int, long, double, string |
Use the value stored in an SDF property in the input molecule. The argument is the property name which is mandatory. |
chemterm |
int, long, double, string |
Execute a Chemical Terms expression evaluation on the input molecule. The argument is the expression to be evaluated which is mandatory. Note that the imported molecule will be passed for Chemical Terms evaluation WITHOUT SDF properties. Please note that you might need additional licenses for executiong certain calculations. |
Example
The following invocation is based on the example script rest-api-example.sh (see Self contained exmaples for details) where various SDF properties are extracted and stored:
bin/calculateOverlap.sh \
-name pubchem1k-in-drugbank-cfp7 \
-descname cfp7 \
-context createSimpleCfp7Context \
-queryname pubchem1k \
-qmf data/molecules/pubchem-compound/pubchem-compound-rnd-1k.sdf.gz \
-qidname \
-qprop compound_cid:int:sdfprop:PUBCHEM_COMPOUND_CID \
-qprop compound_canonicalized:int:sdfprop:PUBCHEM_COMPOUND_CANONICALIZED \
-qprop cactvs_complexity:double:sdfprop:PUBCHEM_CACTVS_COMPLEXITY \
-qprop cactvs_hbond_acceptor:int:sdfprop:PUBCHEM_CACTVS_HBOND_ACCEPTOR \
-qprop cactvs_hbond_donor:int:sdfprop:PUBCHEM_CACTVS_HBOND_DONOR \
-qprop cactvs_rotatable_bond:int:sdfprop:PUBCHEM_CACTVS_ROTATABLE_BOND \
-qprop exact_mass:double:sdfprop:PUBCHEM_EXACT_MASS \
-qprop molecular_weight:double:sdfprop:PUBCHEM_MOLECULAR_WEIGHT \
-qprop cactvs_tpsa:double:sdfprop:PUBCHEM_CACTVS_TPSA \
-qprop monoisotopic_weight:double:sdfprop:PUBCHEM_MONOISOTOPIC_WEIGHT \
-qprop total_charge:int:sdfprop:PUBCHEM_TOTAL_CHARGE \
-qprop heavy_atom_count:int:sdfprop:PUBCHEM_HEAVY_ATOM_COUNT \
-qprop atom_def_stereo_count:int:sdfprop:PUBCHEM_ATOM_DEF_STEREO_COUNT \
-qprop atom_udef_stereo_count:int:sdfprop:PUBCHEM_ATOM_UDEF_STEREO_COUNT \
-qprop bond_def_stereo_count:int:sdfprop:PUBCHEM_BOND_DEF_STEREO_COUNT \
-qprop bond_udef_stereo_count:int:sdfprop:PUBCHEM_BOND_UDEF_STEREO_COUNT \
-qprop isotopic_atom_count:int:sdfprop:PUBCHEM_ISOTOPIC_ATOM_COUNT \
-qprop cactvs_tauto_count:int:sdfprop:PUBCHEM_CACTVS_TAUTO_COUNT \
-targetname drugbank \
-tmf data/molecules/drugbank/drugbank-common_name.smi.gz \
-tidname \
-tprop chemterms_atom_count:int:chemterm:atomCount \
-tprop chemterms_rotatable_bonds:int:chemterm:rotatableBondCount \
-tprop chemterms_ring_count:int:chemterm:ringCount \
-tprop chemterms_mass:double:chemterm:mass \
-out pubchem1k-drugbank-overlap.bin
Execution time is a few seconds. Launching the embedded server:
bin/gui.sh -in pubchem1k-drugbank-overlap.bin -port 8085
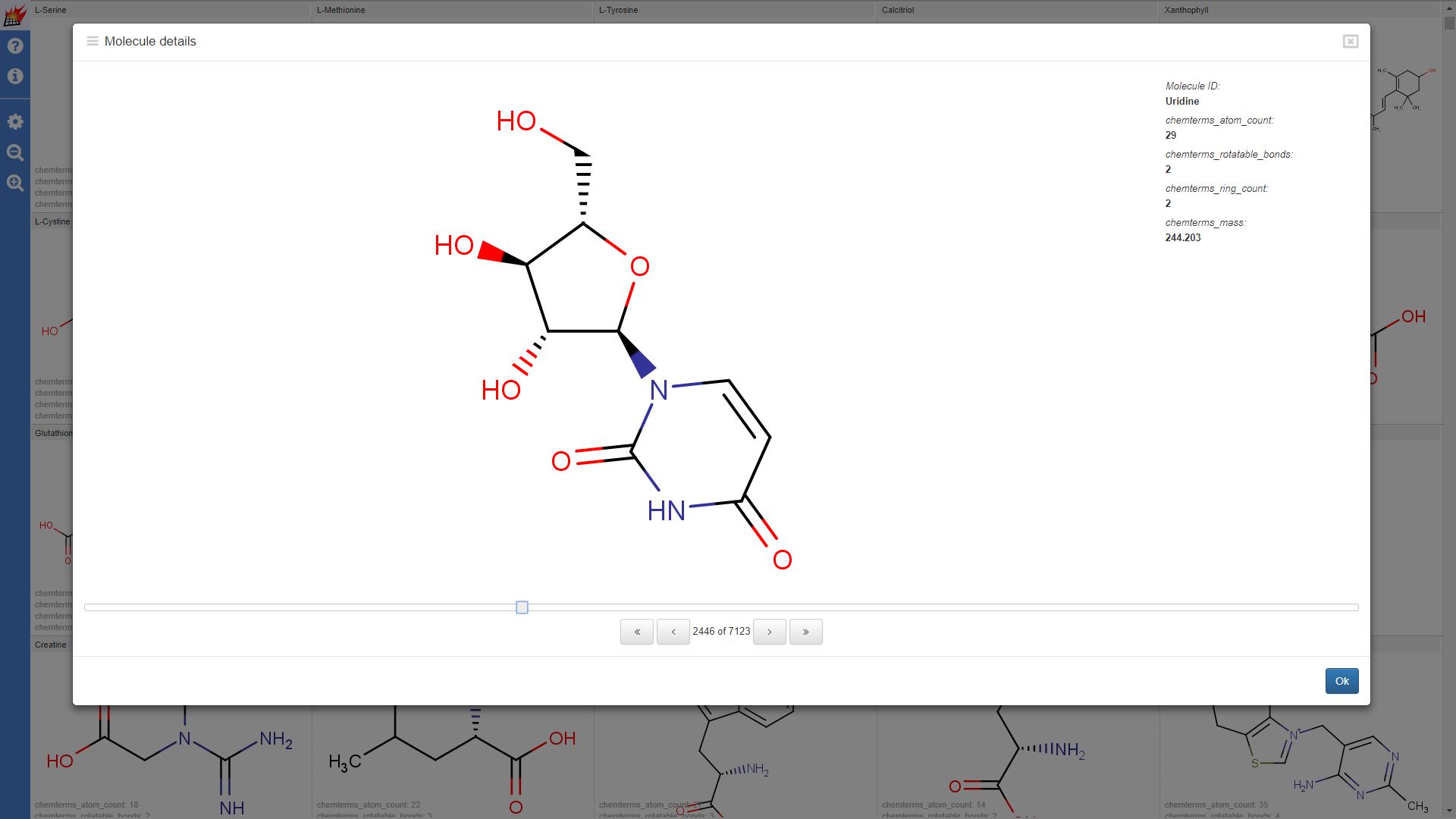
Display additional data
Properties are shown in the molecule table view and molecule details dialog of the pubchem1k molecule set (Note that "hydrogene display" setting was set to "Display dehydrogenized structures" by default; cell size for the molecule table was increased):
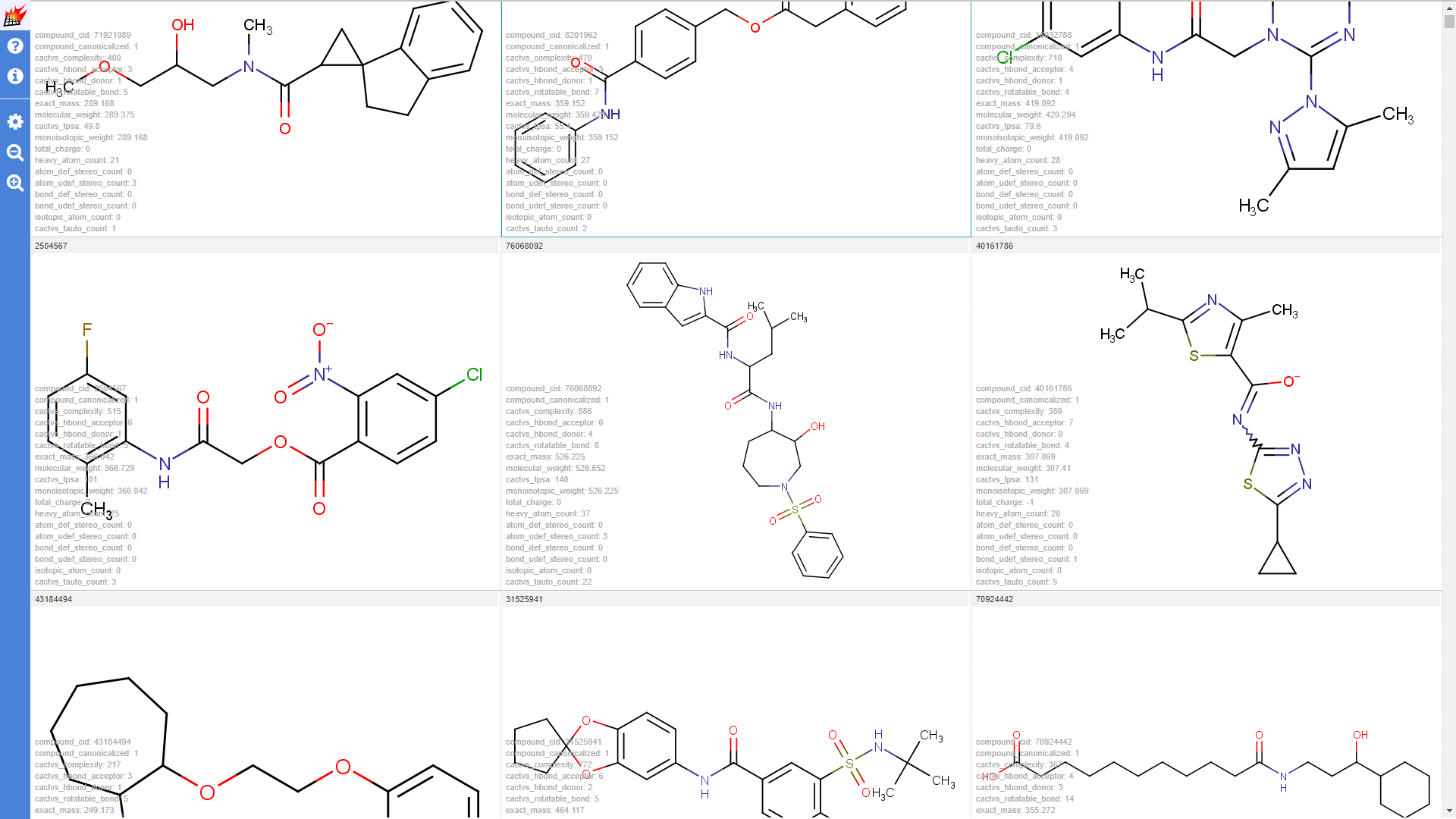
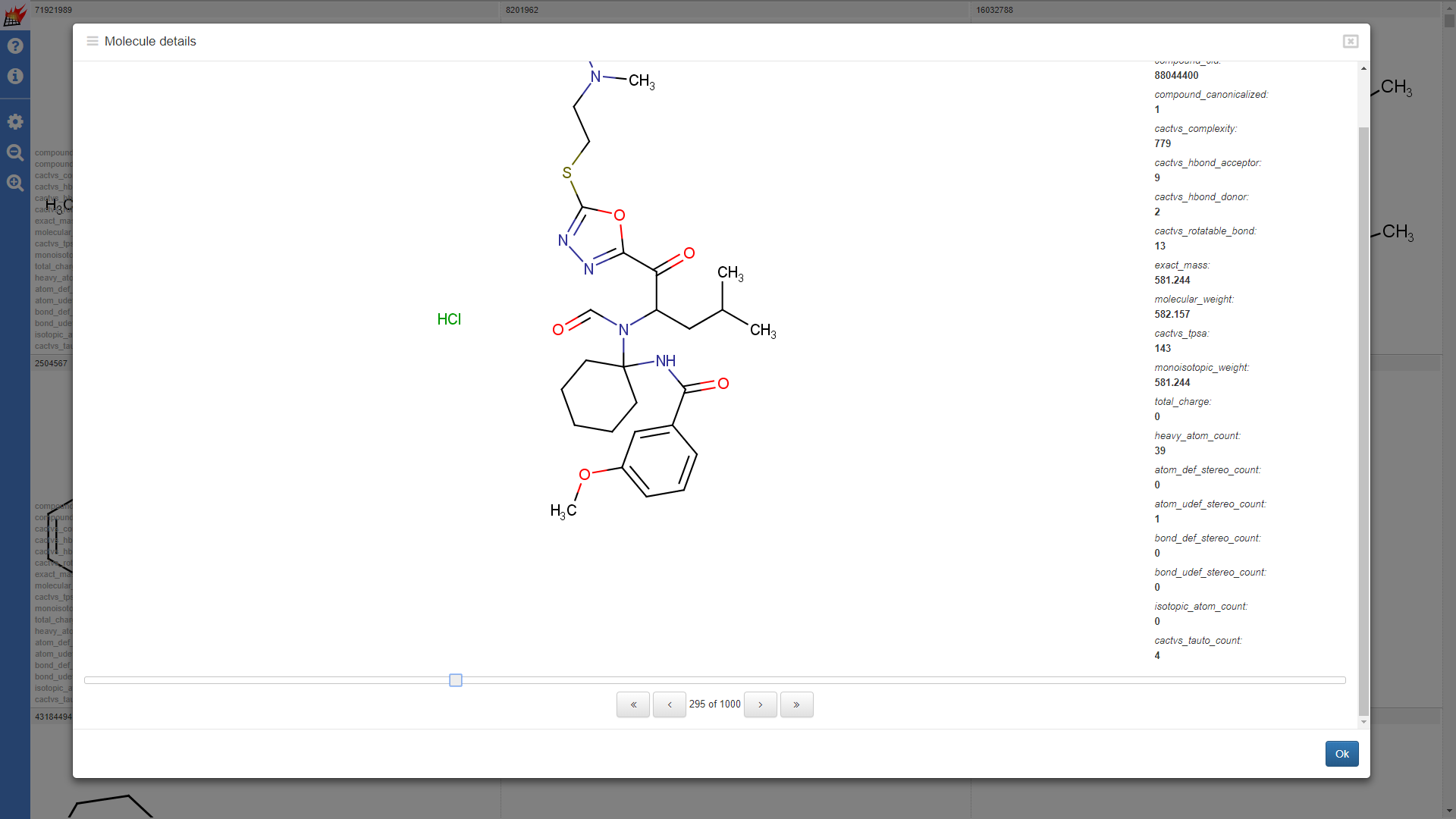
Properties shown for the drugbank-1 dataset:
Use additional data in overlap analysis visualization
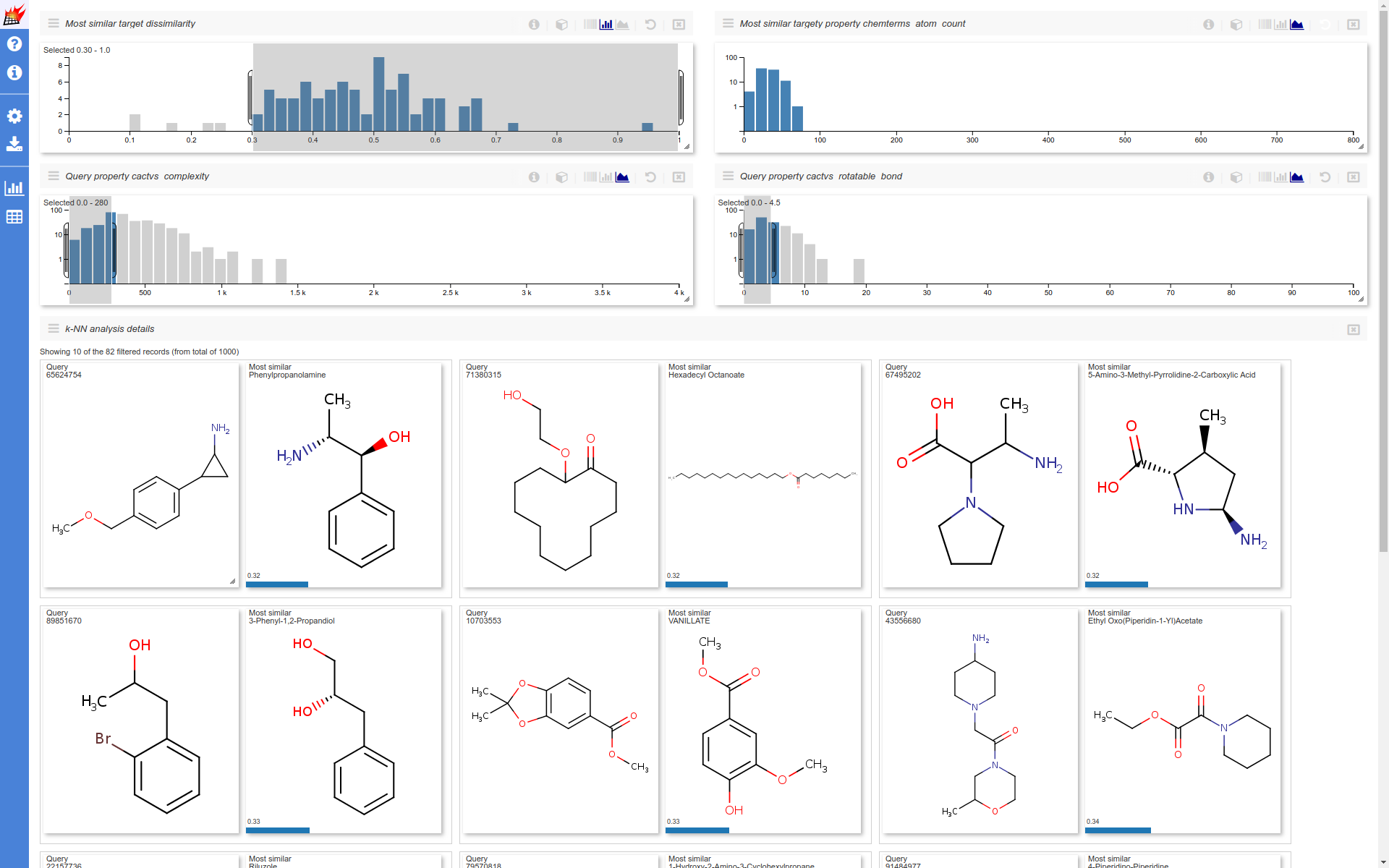
Distribution of numeric stored additional data can also be selected as histogram dimension. In the following example the non overlapping queries (for which no similar target was found) are selected. The selection is further narrowed for queries having small cactvs_complexity and cactvs_rotatable_bond (coming from PubChem) values. Distibution of the remaining most similar targets atom_count (calculated by Chemical Terms) are also shown.
(Note that "hydrogenbe display" setting was set to "Display dehydrogenized structures"):
Use k-NN analysis for property space exploration
The current visualization capabilities of the k-NN analysis can be used for single molecule set property space analysis. For this demonstration we will use the shipped chembl-21 dataset as a query set with properties calculated by chemical terms. As a target set we will use a single molecule containing a single carbon atom:
bin/calculateOverlap.sh \
-name propspace-of-chembl-cfp7 \
-descname cfp7 \
-context createSimpleCfp7Context \
-queryname chembl \
-qmf data/molecules/chembl/chembl-21.smi.gz \
-qidname \
-qprop chemterms_atom_count:int:chemterm:atomCount \
-qprop chemterms_rotatable_bonds:int:chemterm:rotatableBondCount \
-qprop chemterms_ring_count:int:chemterm:ringCount \
-qprop chemterms_mass:double:chemterm:mass \
-targetname carbon \
-tm "C a carbon" \
-tidname \
-out propspace-of-chembl-overlap.bin
Execution time (on an i7-4790 desktop machine) is around 4 minutes.
bin/gui.sh -in propspace-of-chembl-overlap.bin -port 8085
This analysis as well a similar one on the nci-250k dataset is included in self contained example script examples/overlap-example.sh.
After opening the k-NN visualization page the following changes are made to the page layout
- Four additional histograms are added
- The displayed histograms are set to show the individual query properties and the query indices
- Histogram y axes are set to logarithmic (with the exception of the query indices)
- The molecule table is set to show
0most similar targets (only the selected queries shown) and to show4records. - Molecule table cell size is increased
- Components are reordered - molecule table on the left, histograms on the right
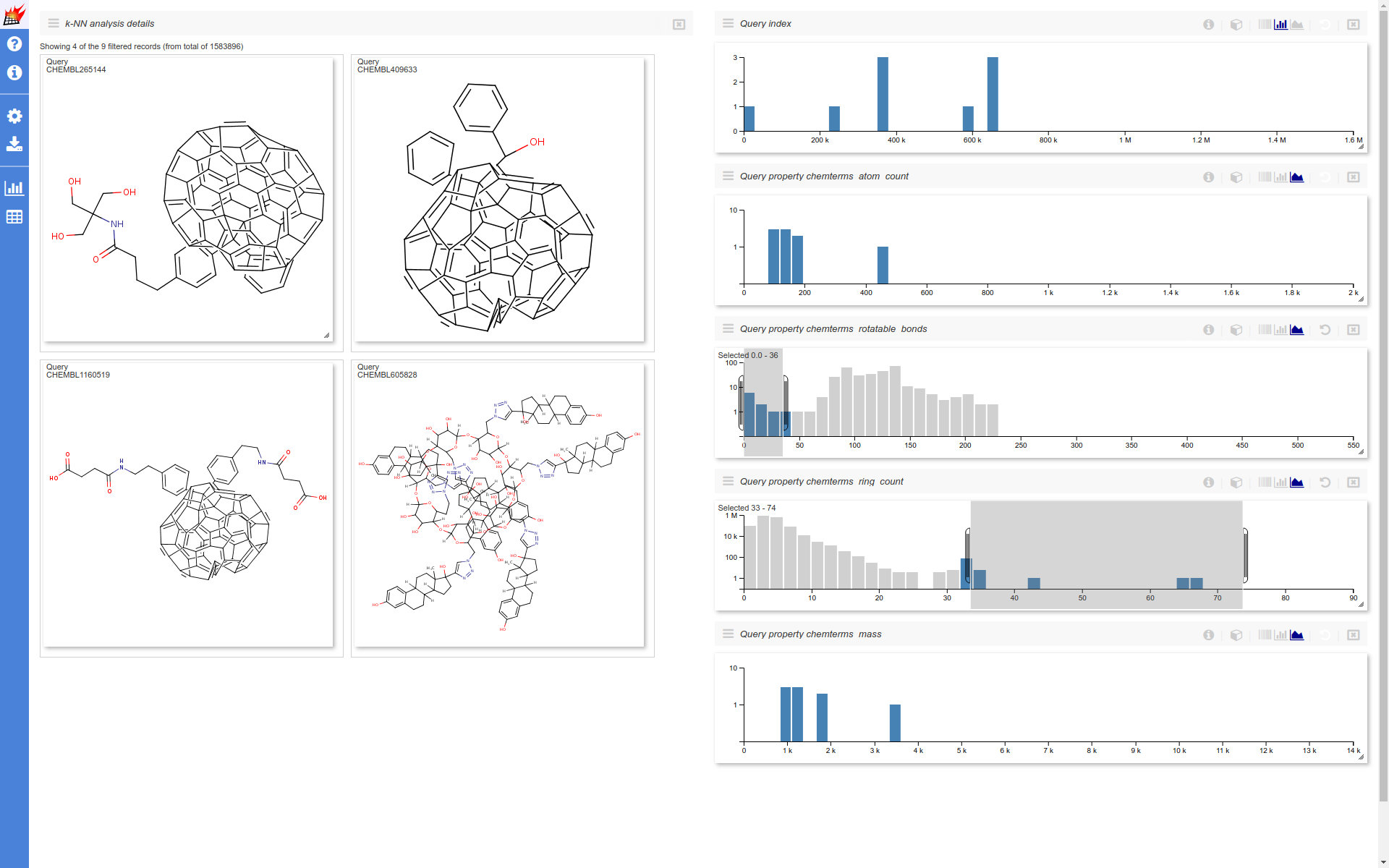
This example visualization handles around nearly 8M data points on the client (browser) side (~1.6 M molecules * 5 displayed dimensions). The response time of the UI for certain interactions falls into the 1-2 seconds range. This is expected to be improved in the further versions.
Accessing additional data through REST API
See REST API endpoint molecules for detailed documentation. A few examples using curl are given. To use these examples launch examples/rest-api-example.sh script.
Metadata on stored properties
curl -g "http://localhost:8085/rest/molecules/pubchem1k" | python -m json.tool
{
"absentids": 0,
"absentmols": 0,
"description": "pubchem1k (from pubchem1k-drugbank-overlap.bin)",
"name": "pubchem1k",
"propnames": [
"compound_cid",
"compound_canonicalized",
"cactvs_complexity",
"cactvs_hbond_acceptor",
"cactvs_hbond_donor",
"cactvs_rotatable_bond",
"exact_mass",
"molecular_weight",
"cactvs_tpsa",
"monoisotopic_weight",
"total_charge",
"heavy_atom_count",
"atom_def_stereo_count",
"atom_udef_stereo_count",
"bond_def_stereo_count",
"bond_udef_stereo_count",
"isotopic_atom_count",
"cactvs_tauto_count"
],
"props": [
{
"extractor": "Get SDF property \"PUBCHEM_COMPOUND_CID\" as java.lang.Integer. Missing values allowed: false",
"name": "compound_cid",
"numeric": true,
"type": "java.lang.Integer"
},
{
"extractor": "Get SDF property \"PUBCHEM_COMPOUND_CANONICALIZED\" as java.lang.Integer. Missing values allowed: false",
"name": "compound_canonicalized",
"numeric": true,
"type": "java.lang.Integer"
},
...
{
...
}
],
"size": 1000,
"url": "rest/molecules/pubchem1k"
}
Single property on a single structure
curl -g "http://localhost:8085/rest/molecules/pubchem1k/10/props/compound_cid" | python -m json.tool
{
"value": 72819133
}
Properties for multiple molecules (index range)
curl -X POST \
-H "Content-Type: application/x-www-form-urlencoded" \
-d 'start=10' \
-d 'maxcount=20' \
-g "http://localhost:8085/rest/molecules/pubchem1k/props/compound_cid/get-properties-on-index-range" | python -m json.tool
{
"count": 20,
"start": 10,
"values": [
72819133,
26175566,
66777683,
43934213,
68924857,
73745083,
81901454,
6127448,
52060933,
23288074,
66710358,
68665148,
76605832,
73377643,
60711711,
64497837,
62627366,
31893941,
20771615,
81785057
]
}
Multiple properties for multiple molecules
curl -X POST \
-H "Content-Type: application/x-www-form-urlencoded" \
-d 'indices[]=10&indices[]=11&indices=12&indices[]=20' \
-d 'props[]=compound_cid&props[]=molecular_weight' \
-g "http://localhost:8085/rest/molecules/pubchem1k/get-multiple-props" | python -m json.tool
{
"props": {
"compound_cid": {
"10": 72819133,
"11": 26175566,
"20": 66710358
},
"molecular_weight": {
"10": 940.668,
"11": 460.951,
"20": 169.26
}
}
}