Introduction to overlap analysis
Similarity based overlap analysis is based on exhaustive k nearest neighbor search. For every structure from the input set k of its nearest neighbors (most similar target structures) are fetched from the target set. Tool calculateOverlap.sh can read the query and target molecule sets, calculate fingerprints, invoke the similarity based overlap analysis calculation and store all imported/calculated data in a single binary file. This binary file can be read by the embedded server gui.sh and exposed to the Web UI for interactive visualization.
Invoke a basic comparison
To invoke the analysis using the sample datasets launch
bin/calculateOverlap.sh \
-context createSimpleCfp7Context \
-qmf data/molecules/antibiotics/antibiotics.smi.gz \
-qidname \
-tmf data/molecules/who-essential-medicines/who-essential-medicines.smi.gz \
-tidname \
-out antibiotics-essentials-overlap.bin
Since the involved sets are small (146 and 342 molecules) the execution is finished in a few seconds.
Breakdown of the invocations
| Command line part | Description |
|---|---|
-context <CONTEXT> |
Specify molecular descriptor, default comparison metric and other parameters to be used during calculation and later search. For details see document Basic overview of the concepts of overlap analysis context. |
-qmf <FILENAME> |
Read queries from molecule file <FILENAME>, parse them and calculate descriptors according the context set. |
-qidname |
Use the molecule name field of the queries as IDs. |
-tmf <FILENAME> |
Read targets from molecule file <FILENAME>, parse them and calculate descriptors according the context set. |
-tidname |
Use the molecule name field of the targets as IDs. |
-out <FILENAME> |
Binary file to write with all the imported and caluclated data. |
To print further help invoke calculateOverlap.sh -h.
Visualization
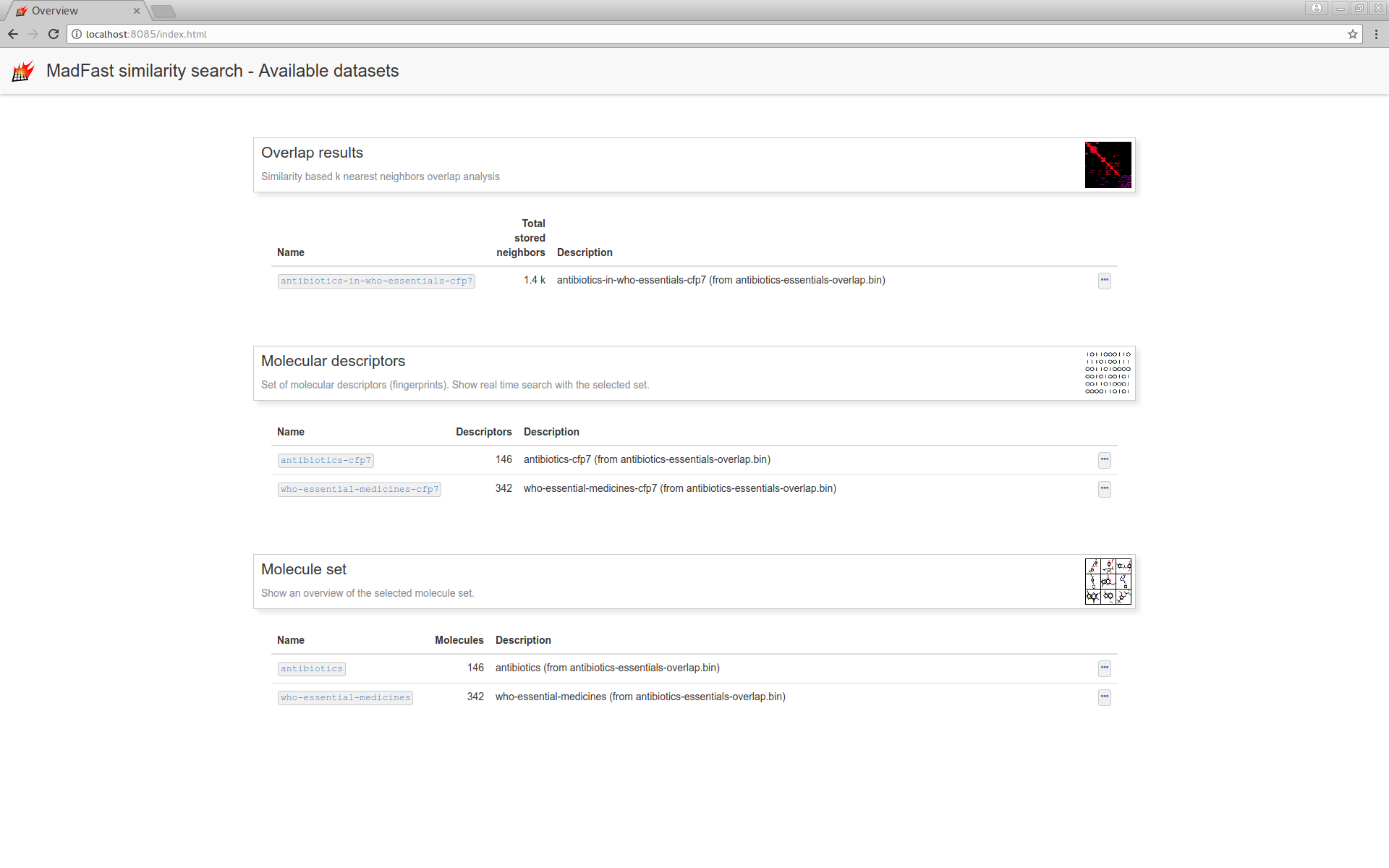
An output file antibiotics-essentials-overlap.bin is created which contains - The query and target molecule sets - One descriptor (fingerprint) calculated for each sets - The search results - The metadata (names) specified as command line options
When this binary file is read by the web server it will expose the read molecules and descriptors as regular resources. The search results are exposed in a form suitable for interactive visualization.
To show visualization simply launch:
bin/gui.sh \
-port 8085 \
-in antibiotics-essentials-overlap.bin
then open http://localhost:8085 in a browser
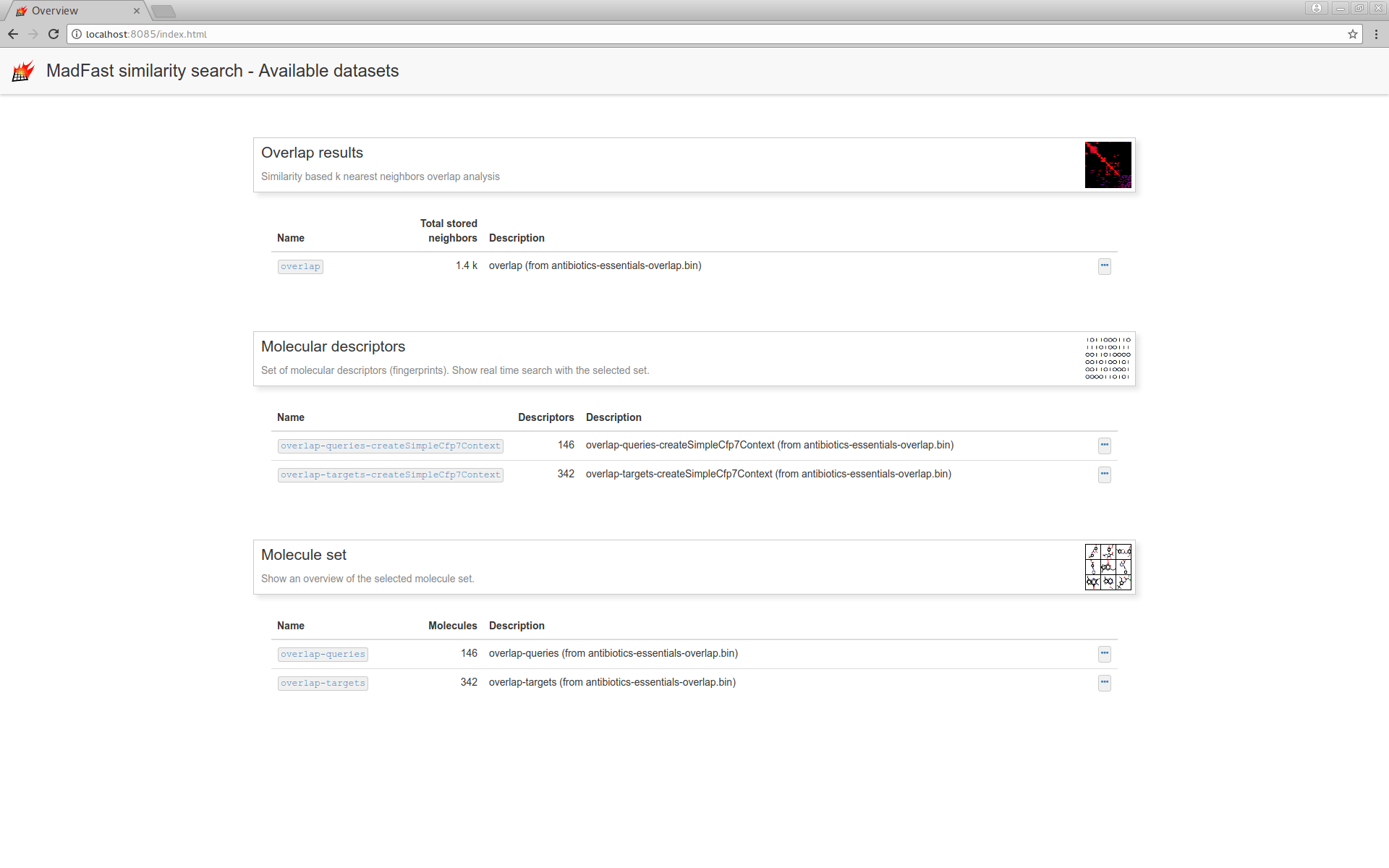
and navigate to the "Overlap results" page:
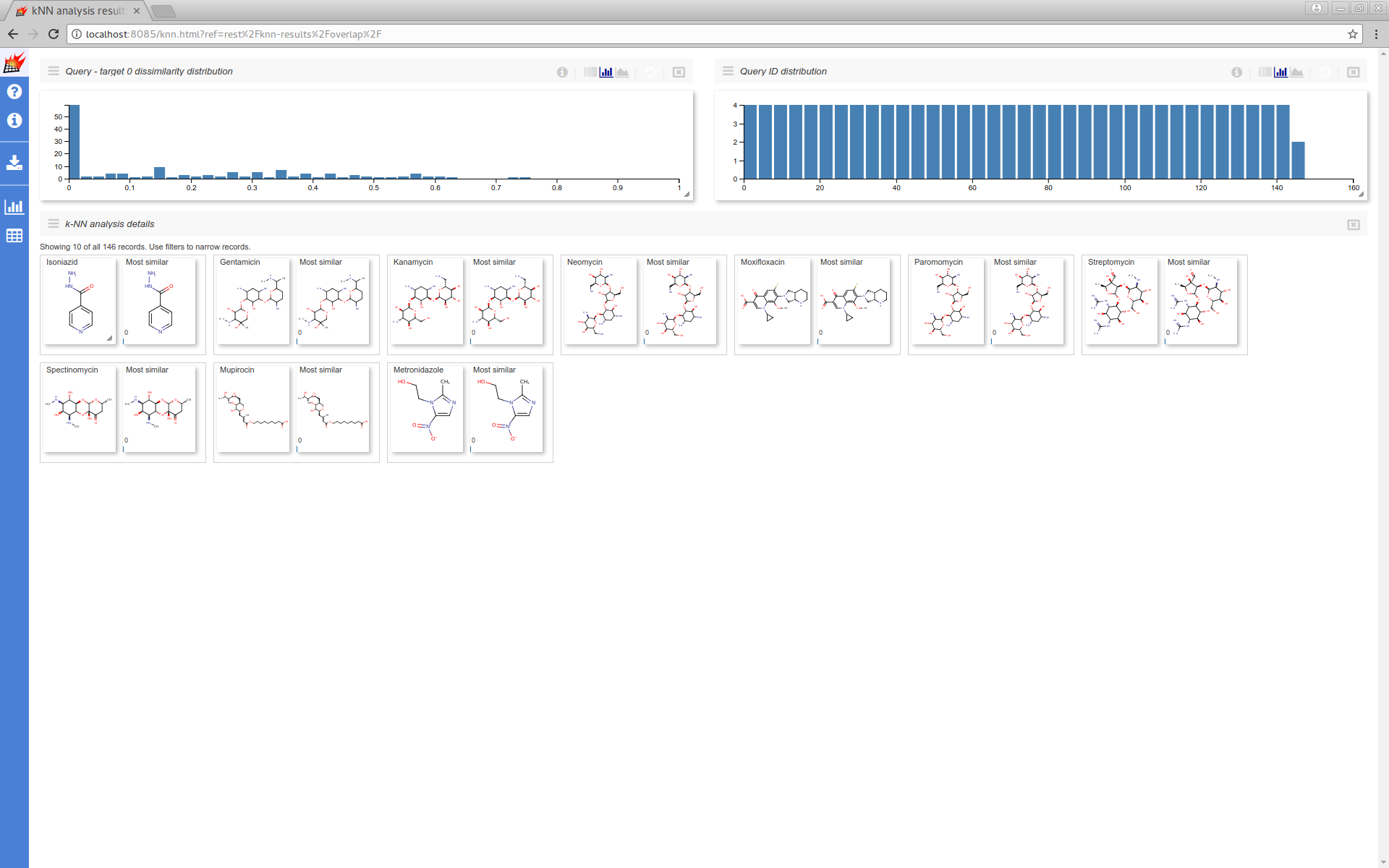
Where you can filter the results dataset and can customize the UI:
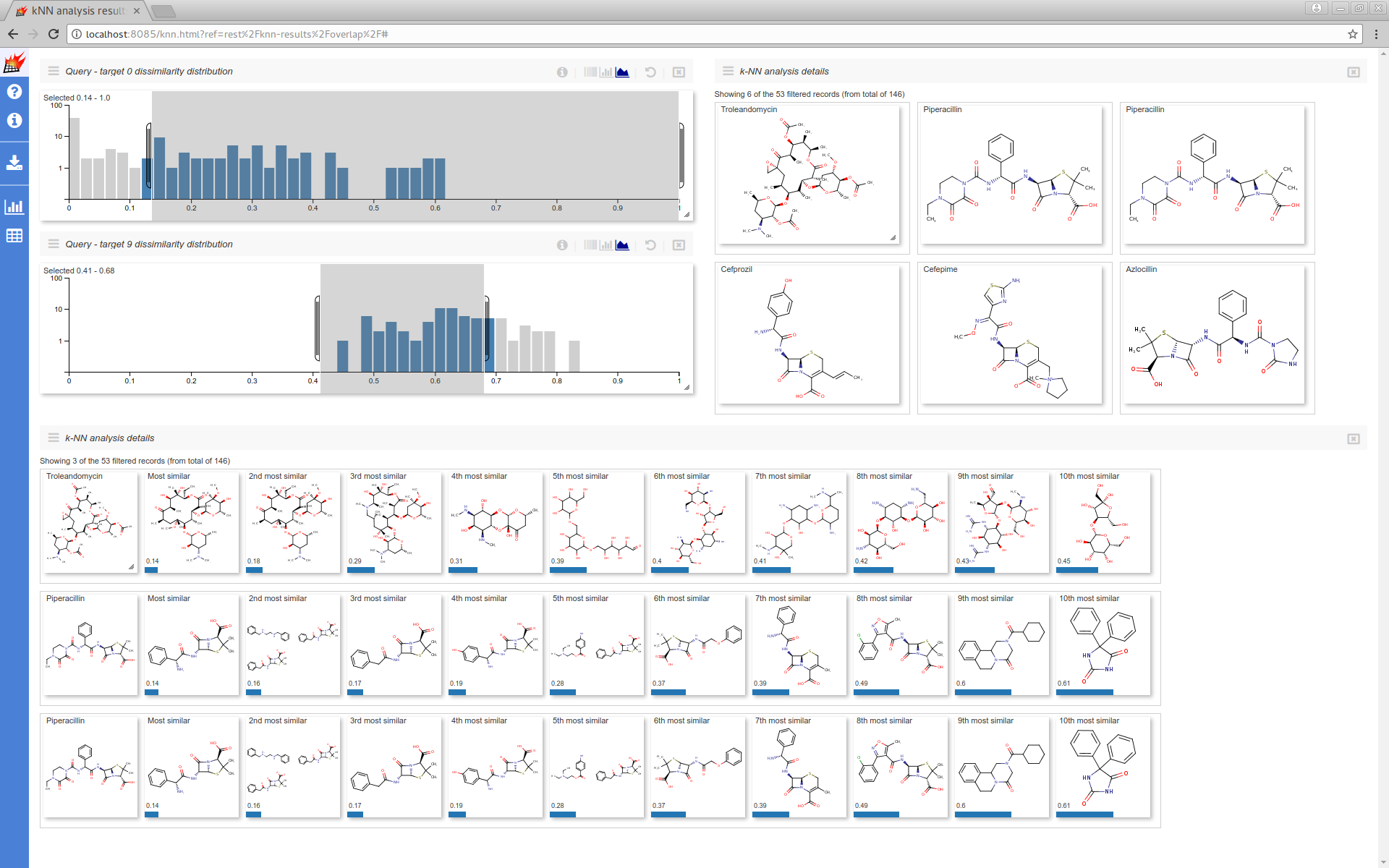
Additional data stored with queries/targets also can be used (see Store additional data for details):
Specify metadata
Descriptions and expected resource names can also be specified:
bin/calculateOverlap.sh \
-name antibiotics-in-who-essentials-cfp7 \
-descname cfp7 \
-context createSimpleCfp7Context \
-queryname antibiotics \
-qmf data/molecules/antibiotics/antibiotics.smi.gz \
-qidname \
-targetname who-essential-medicines \
-tmf data/molecules/who-essential-medicines/who-essential-medicines.smi.gz \
-tidname \
-out antibiotics-essentials-overlap.bin
bin/gui.sh \
-port 8085 \
-in antibiotics-essentials-overlap.bin
Additional properties
Tool calculateOverlap.sh is capable of storing/calculating additional properties for query and target sets. Please see Store additional data for details.
Please note that it is currently possible to use these additional properties in interactive the overlap analysis visualization. Using this additional data as server side search criteria is planned in a future release.
Launch larger analysis
In this example the overlap of the nci-250k dataset (used as a query) with the drugbank dataset (used as a target) is assessed. Also collect profiling and execution statistics data.
bin/calculateOverlap.sh \
-name nci-in-drugbank-cfp7 \
-descname cfp7 \
-context createSimpleCfp7Context \
-queryname nci-250k \
-qmf data/molecules/nci/nci-250k.smi.gz \
-qidname \
-targetname drugbank \
-tmf data/molecules/drugbank/drugbank-common_name.smi.gz \
-tidname \
-out nci-drugbank-overlap.bin \
-stat nci-in-drugbank-stat.txt \
-prof nci-in-drugbank-prof.txt
bin/gui.sh \
-port 8085 \
-in nci-drugbank-overlap.bin \
-profres nci-in-drugbank-prof.txt
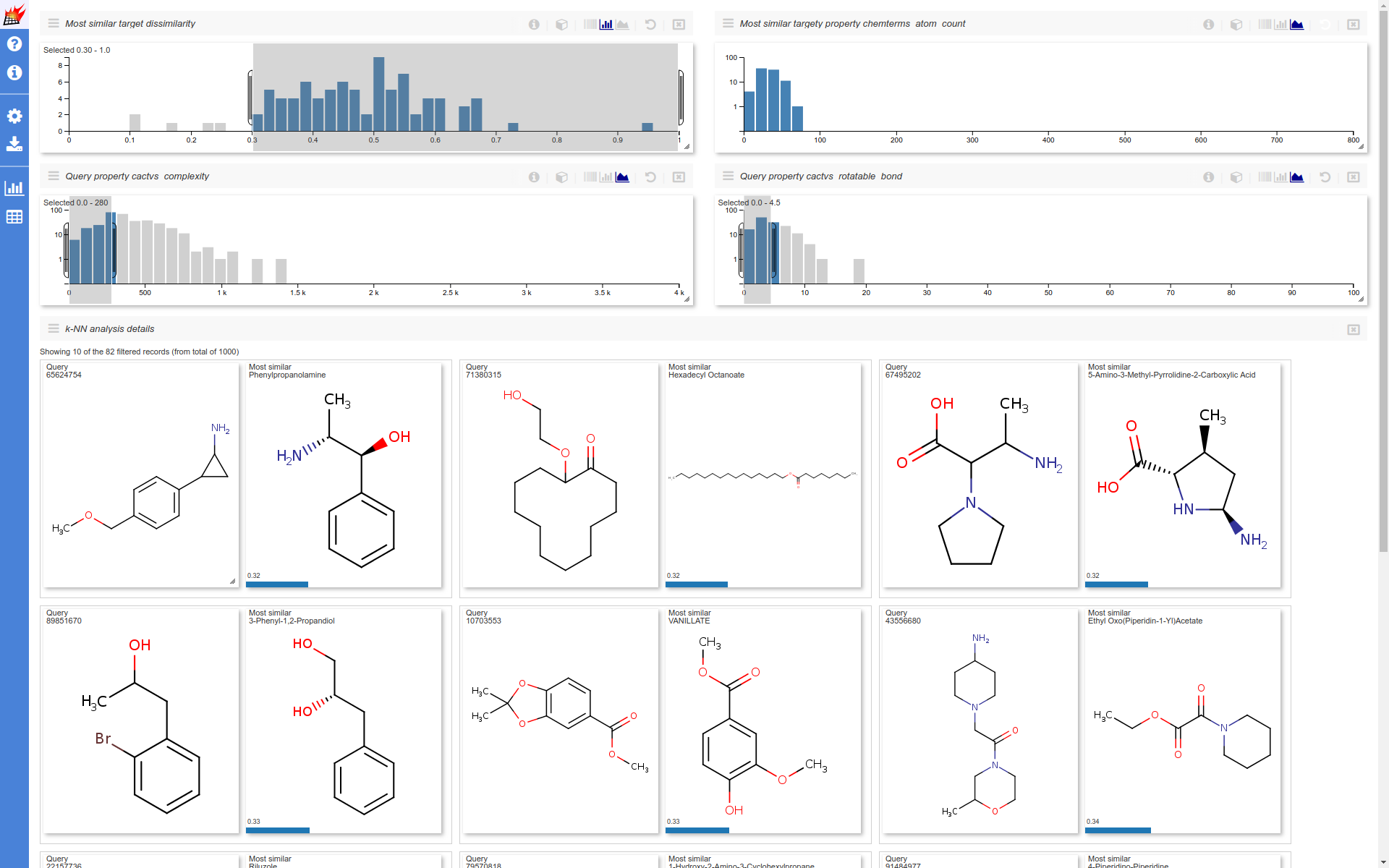
Visualizing some structures from the nci-250k dataset which have the least overlap (most distant nearest neighbor) with the drugbank dataset.
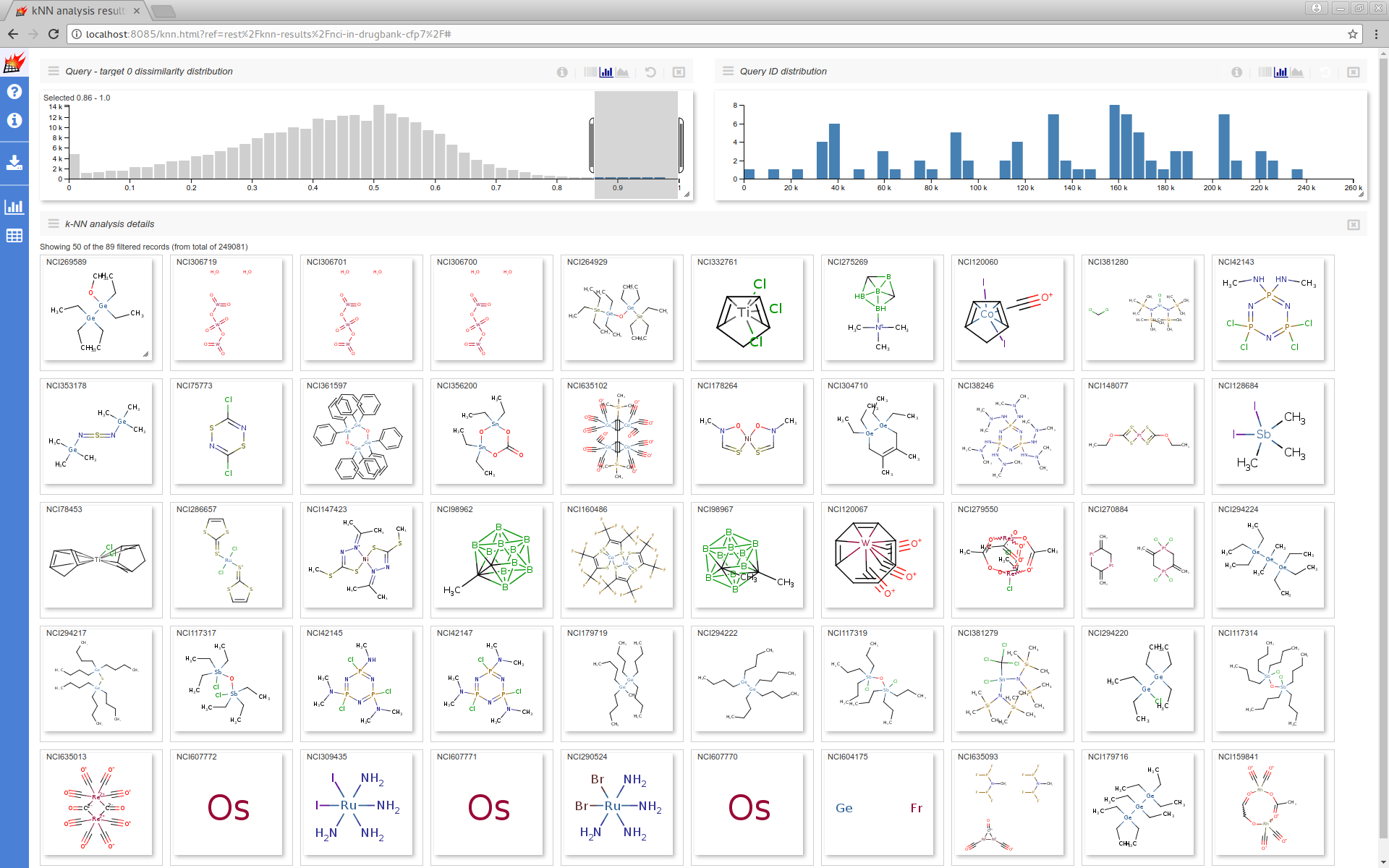
Performance
On an i7 desktop machine the above overlap analysis was done in about 80 seconds.
Resource requirements
The resource requirements of the calculation and the server side of the visualization is composed from the following parts:
- Query and target molecules along with the calculated fingerprints are all stored in memory and in the serialized file. Around 350 MB memory and one minute processing time per million structures are required.
- All the neighbor indices and dissimilarities for each query are stored in the memory, this requires a few ten MB per million neighbors
The client (browser) side of visualization currently fetches data for the shown dimensions (neighbor indices, dissimilarities, additional properties) over the network. This currently usable for up to a few million total data points (query count x used histogram counts).
Limitations
- The shown analysis result dimensions (neighbor indices, dissimilarities and additional properties for each query) are loaded by the Web UI in the browser upon dimension selection. When large number of queries are neighbors are stored this can result in significant memory usage and data transfer. Note that since version 0.3.2 only the data shown on histograms are downloaded.
- When calculating self overlap analysis (query and target sets are the same) the most similar neighbor is usually the query itself. For these use cases the second (and later) most similar neighbors carry meaningful information.
- Histogram binning of overlap data is done in the browser. This scales well for a few hundreds of thousands queries. For more queries (in the multi million range) a server side binning implementation, planned in the future will be required.