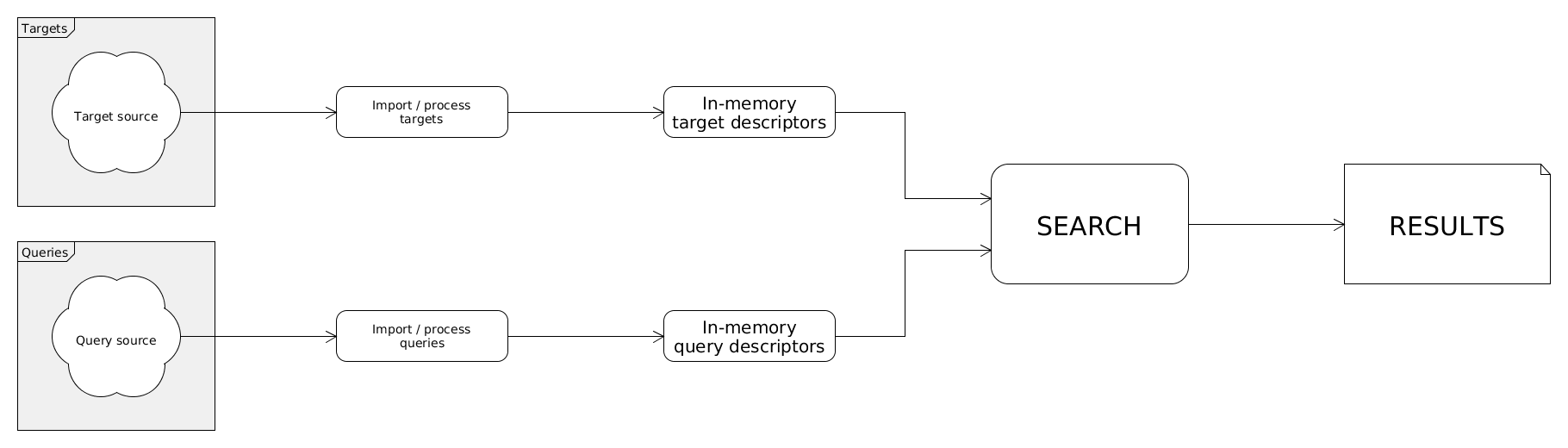
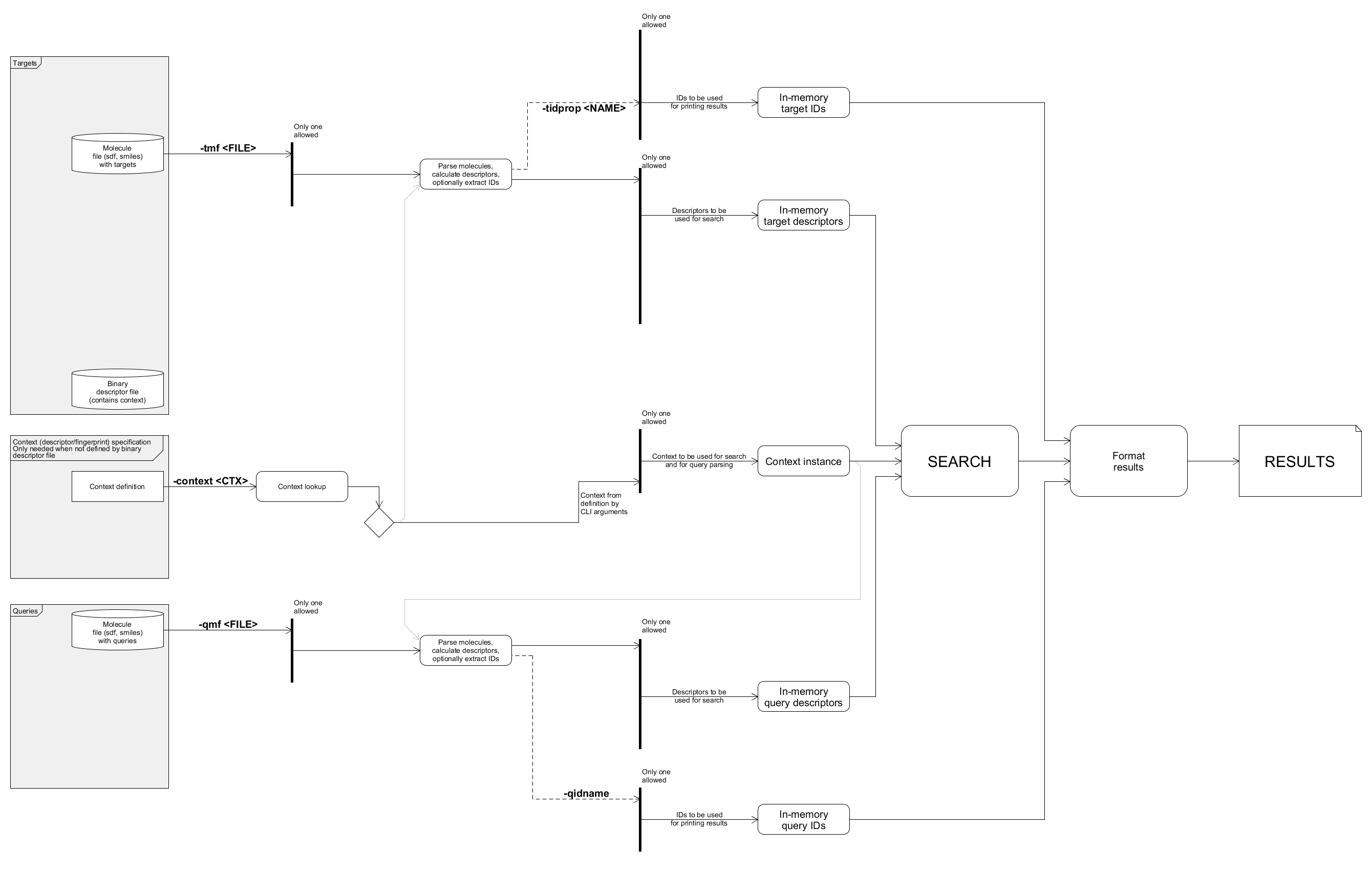
Details on searchStorage
Tool searchStorage provides a command line interface for launching similarity searches. Searching against precomputed descriptors (prepared with buildStorage; detailed in Basic similarity search workflow) is its recommended use case, however on the fly descriptor generation and custom descriptor import is also supported.
High level overview
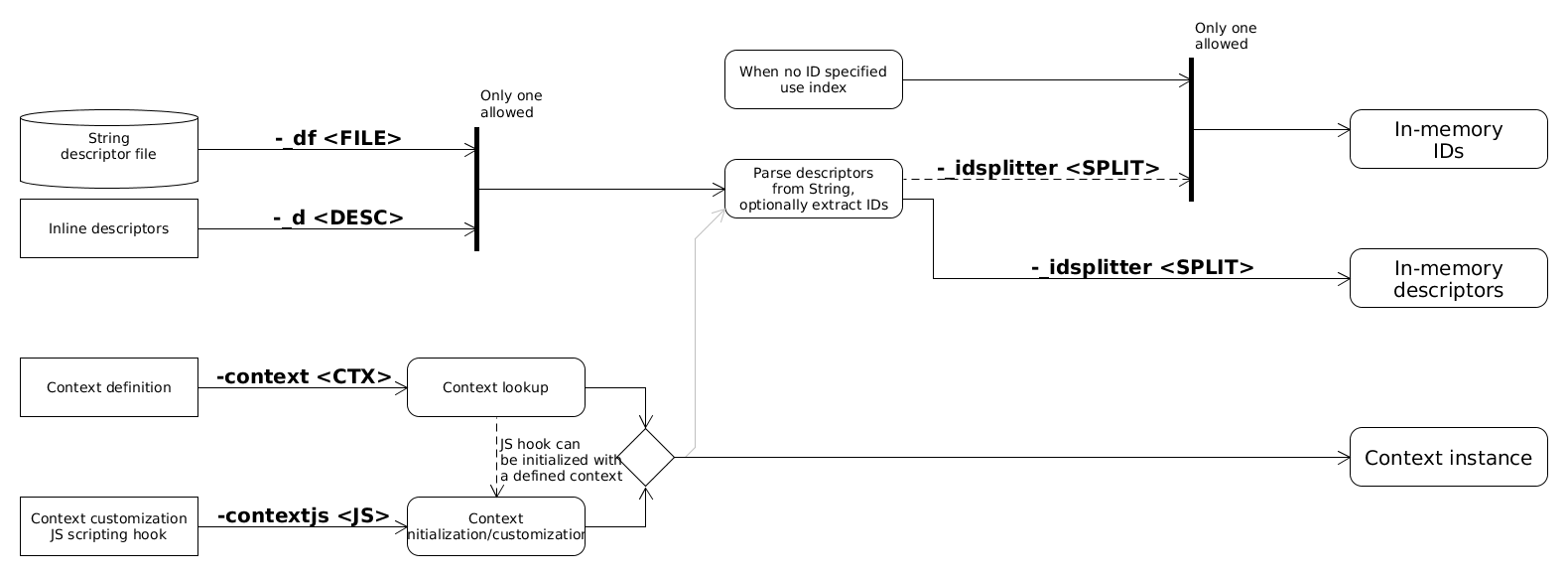
Descriptors can be acquired from various sources. These sources are mutually exclusive; an in-memory descriptor storage can be read from only one source currently. Note that a storage with IDs also can be loaded/imported which will be used for formatting the search results.
Highest level dataflow
The task of searchStorage is to collect query and target descriptors then search queries against targets. Finally it should process search results. The in-memory descriptors can be read/imported from various sources.
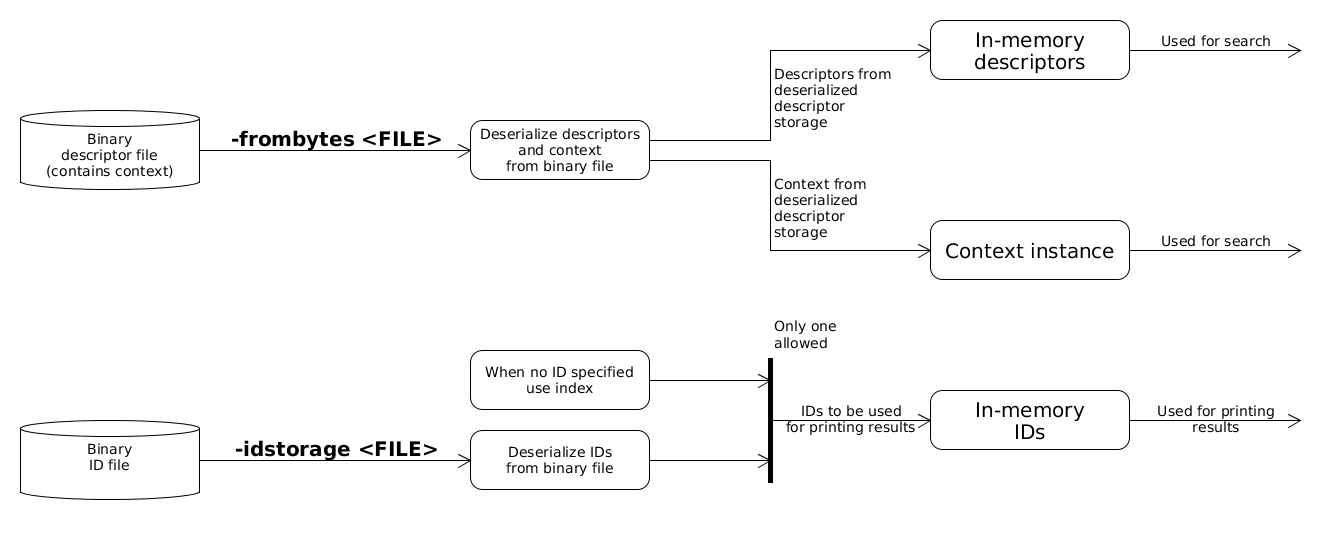
Descriptors from binary file
In-memory descriptors can be deserialized from binary files prepared by buildStorage or importStorage. These binary files store the context used which is needed for search. IDs can also be read from binary files which will be used for printing the results. When ID source is not specified a generated ID storage, representing the indices as IDs will be used. Currently queries can not be read from binary file.
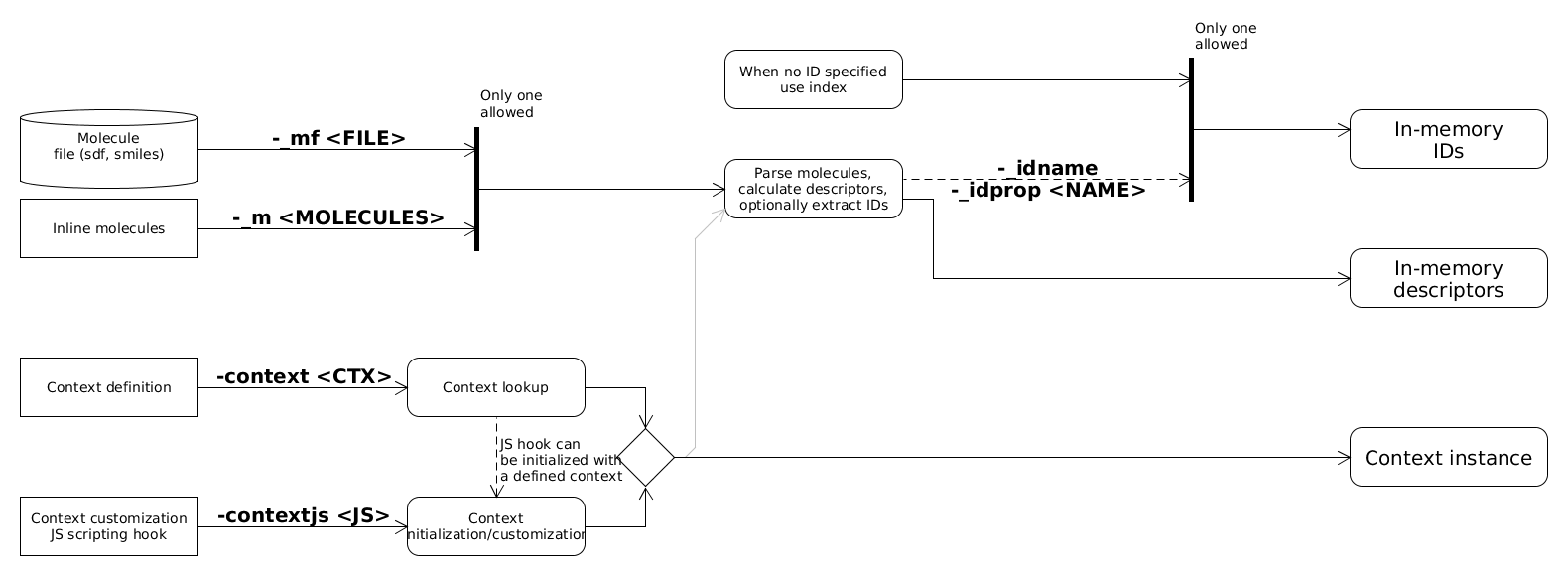
Descriptors calculated for molecules
Molecules can be parsed and descriptors calculated for them. IDs can be extracted from molecule name or an SD property. When no ID is specified a generated ID storage with indices as IDs will be used. To generate descriptors from molecules a context is needed. In this example the context is specified with command line options. Note that when target descriptors are read from binary file the stored context will be used to parse query descriptors. Molecules can be read from a file or they can be specified inline as command line arguments.
Descriptors imported from text
Descriptors can be imported from text source. In this case a context is needed to be specified which describes the textual format. The text source can be stored in an input file or can be specified inline as command line arguments. Each text line is parsed into a descriptor using the specified context. A part of the input text line can be used as IDs when an ID splitter is specified.
Detailed data flow
The following diagram gives an overview of the tools internal data flow composed from the details above as well as relevant command line options.
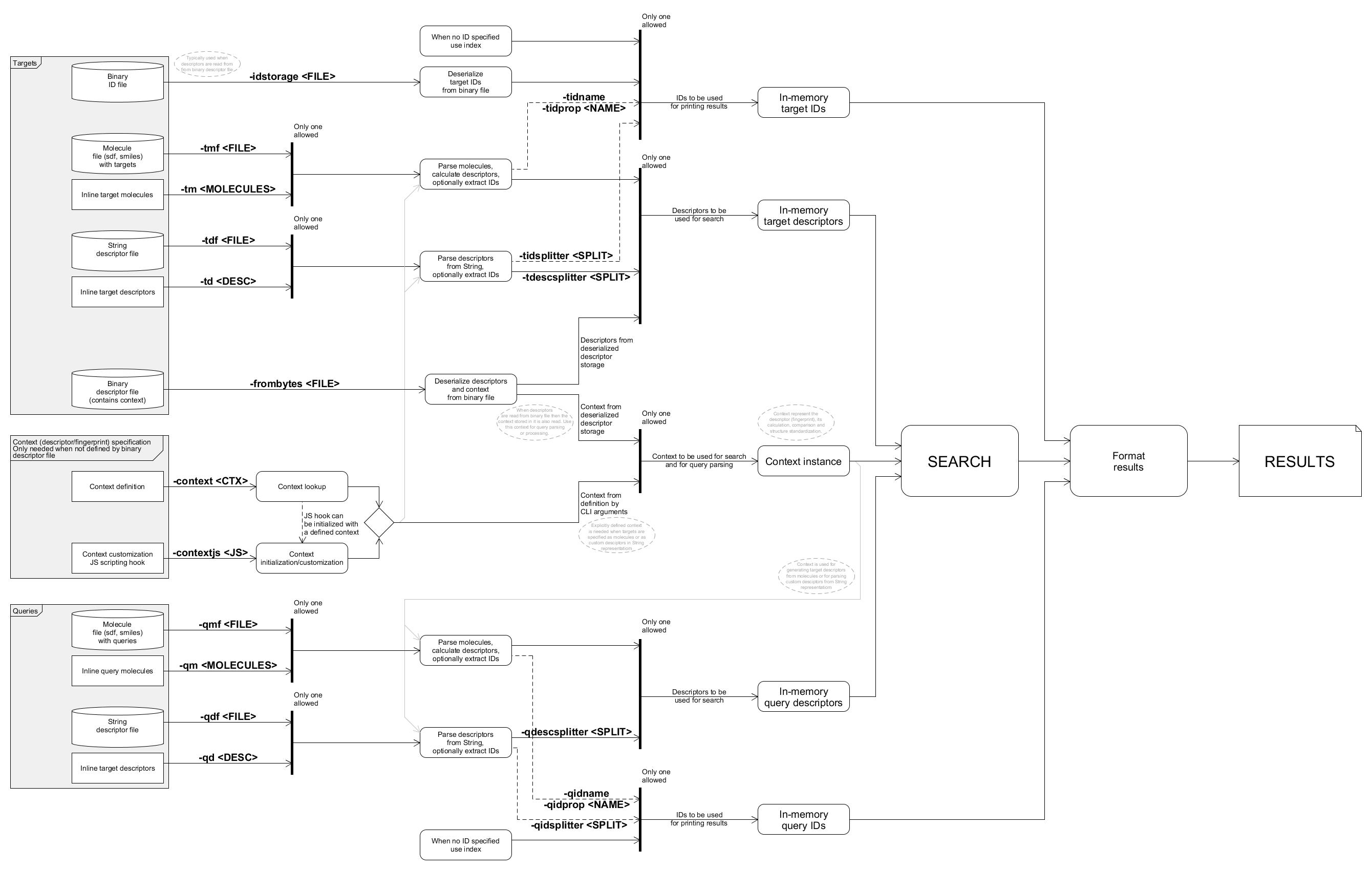
This chart shows the main data paths for collecting the queries, targets and their IDs for searches. Command line options relevant for each data path segments are marked. Note that query and target IDs are always used for printing results, however when they are not specified simply the query / target indices will be used.
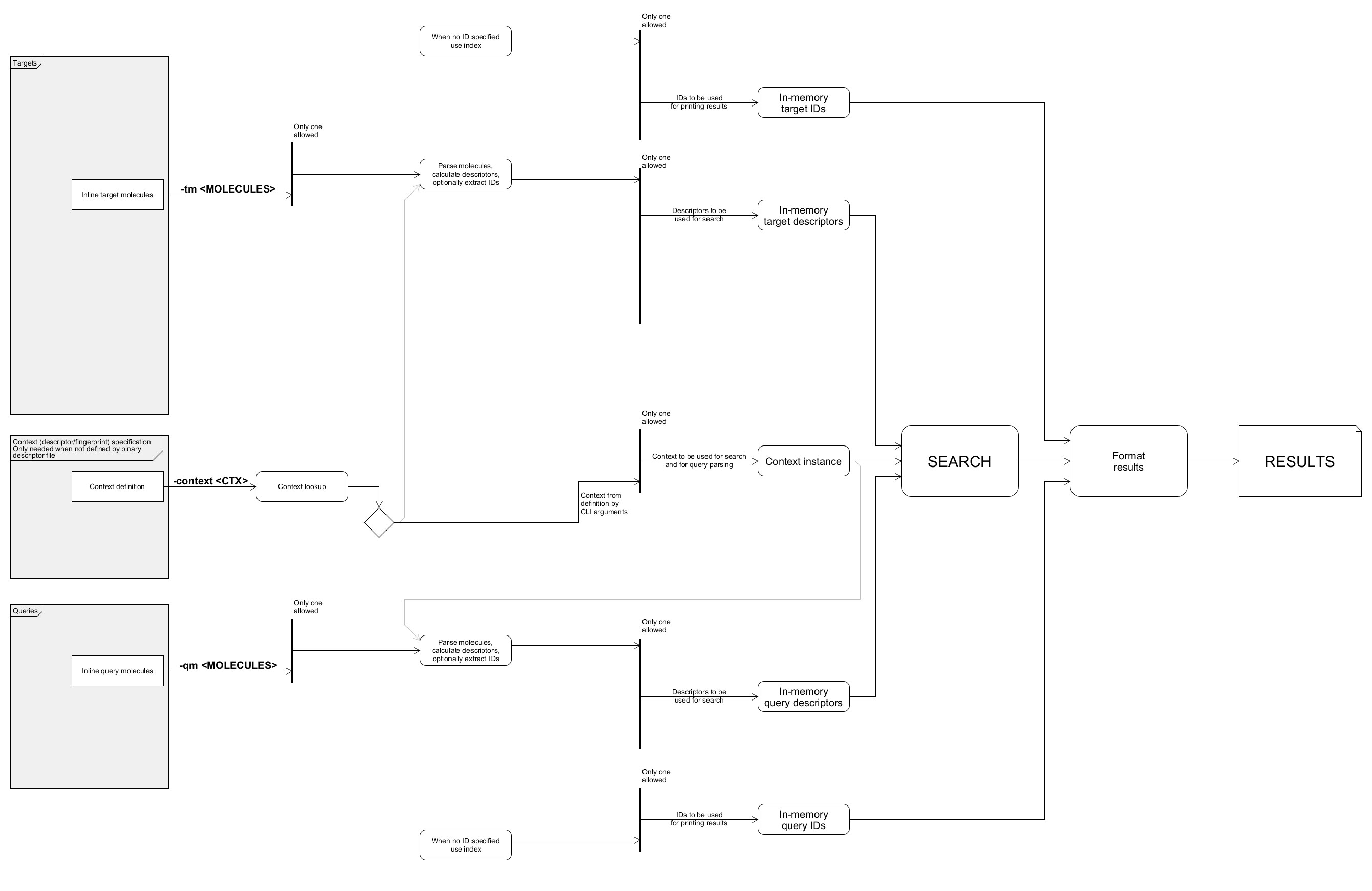
Example: comparing inline molecules
The following command compares molecules specified as inline arguments:
bin/searchStorage.sh \
-context createSimpleCfp7Context \
-qm "C1CCCCC1 cyclohexane" \
-tm "C1CCCCC1CC ethylcyclohexane"
The output:
Query Target Dissimilarity
0 0 0.2
Data flow of this example with unused paths removed:
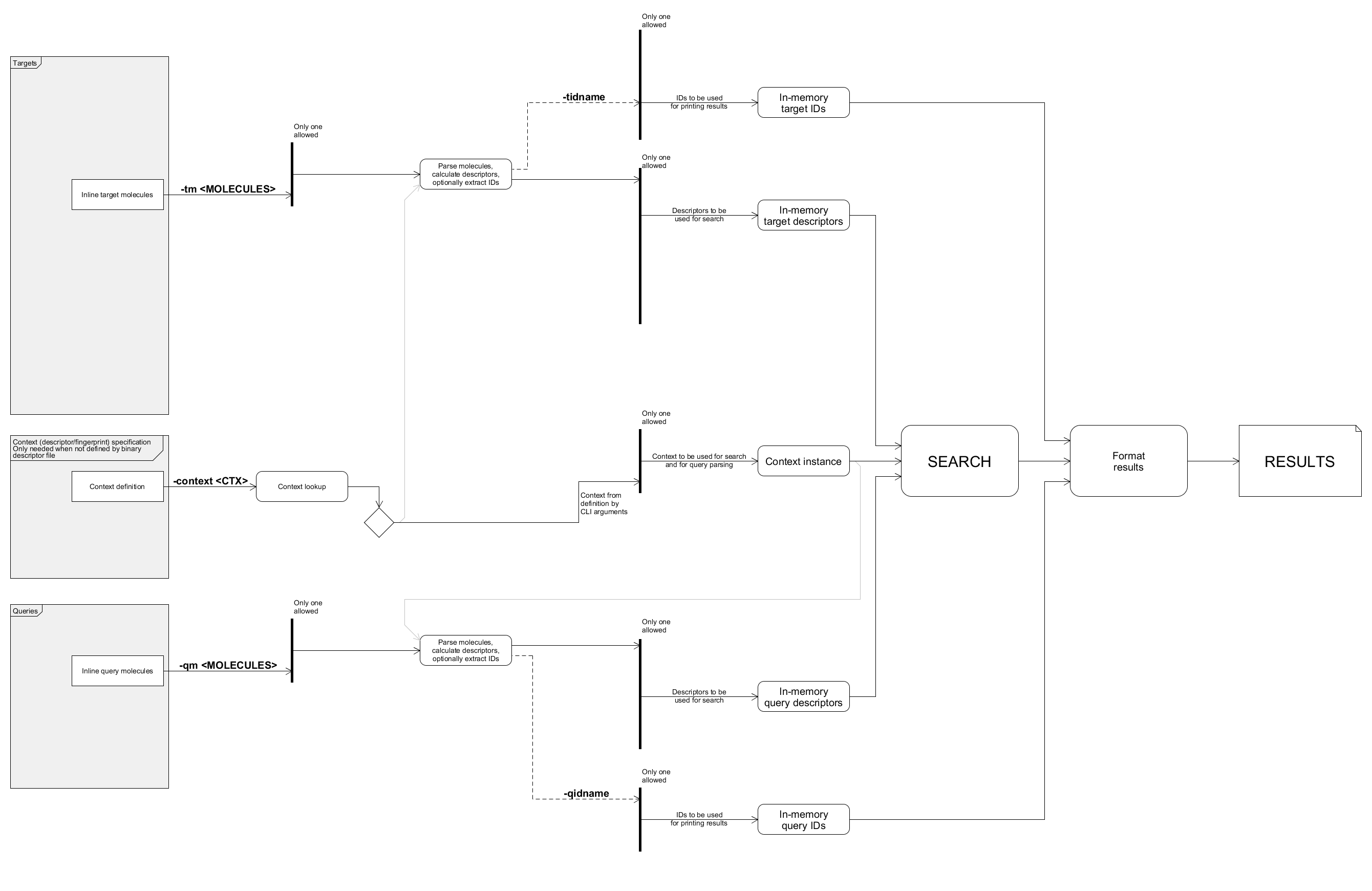
Example: comparing inline molecules with IDs
IDs can be imported and printed. Note that tab size of the terminal is adjusted:
tabs 20
bin/searchStorage.sh \
-context createSimpleCfp7Context \
-qm "C1CCCCC1 cyclohexane" -qidname \
-tm "C1CCCCC1CC ethylcyclohexane" -tidname
The output:
Query Target Dissimilarity
cyclohexane ethylcyclohexane 0.2
Data flow of this example with unused paths removed:
Example: searching against on the fly computed targets
Note that compressed structure files currently not supported, so using gzip to decompress structures and targets are read from stdin. Tab size of the terminal is adjusted.
tabs 40
gzip -dc data/molecules/drugbank/drugbank-all.sdf.gz | bin/searchStorage.sh \
-context createSimpleCfp7Context \
-qmf data/molecules/vitamins/vitamins.smi \
-qidname \
-tmf - \
-tidprop COMMON_NAME
The output:
Query Target Dissimilarity
Vitamin A - Retinol Vitamin A 0.0
Vitamin A - Retinal Alitretinoin 0.14814814814814814
Vitamin A - beta-Carotene 1,3,3-trimethyl-2-[(1E,3E)-3-methylpenta-1,3-dien-1-yl]cyclohexene 0.02631578947368421
Vitamin B1 - Thiamine Thiamine 0.0
Vitamin B2 - Riboflavin Riboflavin 0.0
Vitamin B3 - Niacin Niacin 0.0
Vitamin B3 - Nicotinamide Nicotinamide 0.0
Vitamin B5 - Pantothenic acid Pantothenic acid 0.0
Vitamin B6 - Pyridoxine Pyridoxine 0.0
Vitamin B6 - Pyridoxal Pyridoxal 0.0
Vitamin B7 - Biotin Biotin 0.0
Vitamin B9 - Folic acid Folic Acid 0.0
Vitamin B9 - Folinic acid Leucovorin 0.0
Vitamin B12 - Cyanocobalamin Hydroxocobalamin 0.10152284263959391
Vitamin B12 - Hydroxocobalamin Hydroxocobalamin 0.08740359897172237
Vitamin B12 - Methylcobalamin Hydroxocobalamin 0.08740359897172237
Vitamin C - Ascorbic acid Vitamin C 0.0
Vitamin D3 - Cholecalciferol Cholecalciferol 0.0
Vitamin D3 - Ergocalciferol Ergocalciferol 0.0
Vitamin E - alpha-Tocopherol Vitamin E 0.0
Vitamin E - beta-Tocopherol Vitamin E 0.0
Vitamin E - gamma-Tocopherol Vitamin E 0.04424778761061947
Vitamin E - delta-Tocopherol Vitamin E 0.09734513274336283
Vitamin E - alpha-Tocotrienol Vitamin E 0.22627737226277372
Vitamin E - beta-Trocotrienol Vitamin E 0.22627737226277372
Vitamin E - gamma-Trocotrienol Vitamin E 0.26277372262773724
Vitamin E - delta-Trocotrienol Vitamin E 0.30656934306569344
Vitamin K1 - Phylloquinone Phylloquinone 0.0
Vitamin K2 - Menatetrenone Phylloquinone 0.08181818181818182
Vitamin K2 - Menaquinone-7 Phylloquinone 0.08181818181818182
Data flow of this example with unused paths removed:
Search modes
Search mode can be selected by option -mode <MODE>. The following examples use 6 target and 3 query molecules from the vitamins dataset:
head -6 data/molecules/vitamins/vitamins.smi > targets.smi
head -9 data/molecules/vitamins/vitamins.smi | tail -3 > queries.smi
Most similar search
Mode MOSTSIMILAR searches for the most similar target for each query. This is the default search mode. Option -maxdissim <THRESHOLD> limits the maximal dissimilarity returned.
head -6 data/molecules/vitamins/vitamins.smi > targets.smi
head -9 data/molecules/vitamins/vitamins.smi | tail -3 > queries.smi
tabs 35
bin/searchStorage.sh \
-context createSimpleCfp7Context \
-tmf targets.smi \
-tidname \
-qmf queries.smi \
-qidname \
-mode MOSTSIMILAR
Query Target Dissimilarity
Vitamin B3 - Nicotinamide Vitamin B3 - Niacin 0.37037037037037035
Vitamin B5 - Pantothenic acid Vitamin B2 - Riboflavin 0.8652849740932642
Vitamin B6 - Pyridoxine Vitamin B3 - Niacin 0.569620253164557
Most similars search
Mode MOSTSIMILARS searches for a maximum number of the most similar targets for each query. Option -count <COUNT> specifies the maximum number of targets to return for each query. Option -maxdissim <THRESHOLD> limits the maximal dissimilarity returned.
head -6 data/molecules/vitamins/vitamins.smi > targets.smi
head -9 data/molecules/vitamins/vitamins.smi | tail -3 > queries.smi
tabs 35
bin/searchStorage.sh \
-context createSimpleCfp7Context \
-tmf targets.smi \
-tidname \
-qmf queries.smi \
-qidname \
-mode MOSTSIMILARS \
-count 2
Query Target Dissimilarity
Vitamin B3 - Nicotinamide Vitamin B3 - Niacin 0.37037037037037035
Vitamin B3 - Nicotinamide Vitamin B1 - Thiamine 0.8269230769230769
Vitamin B5 - Pantothenic acid Vitamin B2 - Riboflavin 0.8652849740932642
Vitamin B5 - Pantothenic acid Vitamin B1 - Thiamine 0.8823529411764706
Vitamin B6 - Pyridoxine Vitamin B3 - Niacin 0.569620253164557
Vitamin B6 - Pyridoxine Vitamin B1 - Thiamine 0.7976878612716763
Fullmatrix with matrix format
Mode FULLMATRIX returns the result of all query-target comparisons. By default the textual output has a matrix format. In this default mode option -maxdissim <THRESHOLD> is not effective.
head -6 data/molecules/vitamins/vitamins.smi > targets.smi
head -9 data/molecules/vitamins/vitamins.smi | tail -3 > queries.smi
tabs 30
bin/searchStorage.sh \
-context createSimpleCfp7Context \
-tmf targets.smi \
-tidname \
-qmf queries.smi \
-qidname \
-mode FULLMATRIX
Target Query Vitamin B3 - Nicotinamide dissimilarity Query Vitamin B5 - Pantothenic acid dissimilarity Query Vitamin B6 - Pyridoxine dissimilarity
Vitamin A - Retinol 0.9156626506024096 0.8850574712643678 0.9047619047619048
Vitamin A - Retinal 0.8888888888888888 0.8977272727272727 0.9351851851851852
Vitamin A - beta-Carotene 0.9210526315789473 0.927710843373494 0.9405940594059405
Vitamin B1 - Thiamine 0.8269230769230769 0.8823529411764706 0.7976878612716763
Vitamin B2 - Riboflavin 0.8415300546448088 0.8652849740932642 0.8208955223880597
Vitamin B3 - Niacin 0.37037037037037035 0.8953488372093024 0.569620253164557
Fullmatrix with list format
Mode FULLMATRIX can be used together with option -out-matrix-as-list to print a query-target-dissimilarity list similar to other search modes. In this case option -maxdissim <THRESHOLD> can be used to specify a dissimilarity threshold.
head -6 data/molecules/vitamins/vitamins.smi > targets.smi
head -9 data/molecules/vitamins/vitamins.smi | tail -3 > queries.smi
tabs 35
bin/searchStorage.sh \
-context createSimpleCfp7Context \
-tmf targets.smi \
-tidname \
-qmf queries.smi \
-qidname \
-mode FULLMATRIX \
-out-matrix-as-list
Query Target Dissimilarity
Vitamin B3 - Nicotinamide Vitamin A - Retinol 0.9156626506024096
Vitamin B3 - Nicotinamide Vitamin A - Retinal 0.8888888888888888
Vitamin B3 - Nicotinamide Vitamin A - beta-Carotene 0.9210526315789473
Vitamin B3 - Nicotinamide Vitamin B1 - Thiamine 0.8269230769230769
Vitamin B3 - Nicotinamide Vitamin B2 - Riboflavin 0.8415300546448088
Vitamin B3 - Nicotinamide Vitamin B3 - Niacin 0.37037037037037035
Vitamin B5 - Pantothenic acid Vitamin A - Retinol 0.8850574712643678
Vitamin B5 - Pantothenic acid Vitamin A - Retinal 0.8977272727272727
Vitamin B5 - Pantothenic acid Vitamin A - beta-Carotene 0.927710843373494
Vitamin B5 - Pantothenic acid Vitamin B1 - Thiamine 0.8823529411764706
Vitamin B5 - Pantothenic acid Vitamin B2 - Riboflavin 0.8652849740932642
Vitamin B5 - Pantothenic acid Vitamin B3 - Niacin 0.8953488372093024
Vitamin B6 - Pyridoxine Vitamin A - Retinol 0.9047619047619048
Vitamin B6 - Pyridoxine Vitamin A - Retinal 0.9351851851851852
Vitamin B6 - Pyridoxine Vitamin A - beta-Carotene 0.9405940594059405
Vitamin B6 - Pyridoxine Vitamin B1 - Thiamine 0.7976878612716763
Vitamin B6 - Pyridoxine Vitamin B2 - Riboflavin 0.8208955223880597
Vitamin B6 - Pyridoxine Vitamin B3 - Niacin 0.569620253164557
Textual search results
The search results are printed to the standard output which can be redirected to a file using option -out <FILE>. By default the dissimilarity values uses Java Double formatting. Using option -out-format <FORMAT> a custom formatting can be specified which delegates to Java java.text.Format. The following example use %.3f for a fixed 3 digit precision:
# Find the 5 most similar structures using asymmetric tversky with on the fly descriptor calculation
tabs 25
gzip -dc gzip -dc data/molecules/drugbank/drugbank-all.sdf.gz | bin/searchStorage.sh \
-context createSimpleCfp7Context \
-metric "tversky,coeffT:0.01,coeffQ:0.99" \
-tmf - \
-tidprop COMMON_NAME \
-mode MOSTSIMILARS \
-count 5 \
-qm "O1CC1 epoxy" \
-qidname \
-out-numeric-format "%.3f"
The output:
Query Target Dissimilarity
epoxy Sevelamer 0.020
epoxy Colestipol 0.028
epoxy Fosfomycin 0.033
epoxy 3-Oxiran-2ylalanine 0.037
epoxy R-Styrene Oxide 0.040
Heatmap image creation
A heatmap image of the search result can be rendered using option -heatmap-image <IMAGE>. In the generated image a cell is associated with every query-target pair. When a dissimilarity value for a query-target pair is retrieved it will be colored on the map according to the color scale. Image generation works with all (MOSTSIMILAR, MOSTSIMILARS and FULLMATRIX) search modes. The generated image can be customized. For a help on the available customization options invoke bin/searchStorage.sh -h.
Self overlap of the vitamins dataset
bin/searchStorage.sh \
-context createSimpleCfp7Context \
-qmf data/molecules/vitamins/vitamins.smi \
-qidname \
-tmf data/molecules/vitamins/vitamins.smi \
-tidname \
-mode FULLMATRIX \
-out vitamins-fullmatrix.txt \
-heatmap-image vitamins-fullmatrix.png \
-heatmap-image-cellsize 15 \
-heatmap-image-query-ids-length 250 \
-heatmap-image-target-ids-length 250
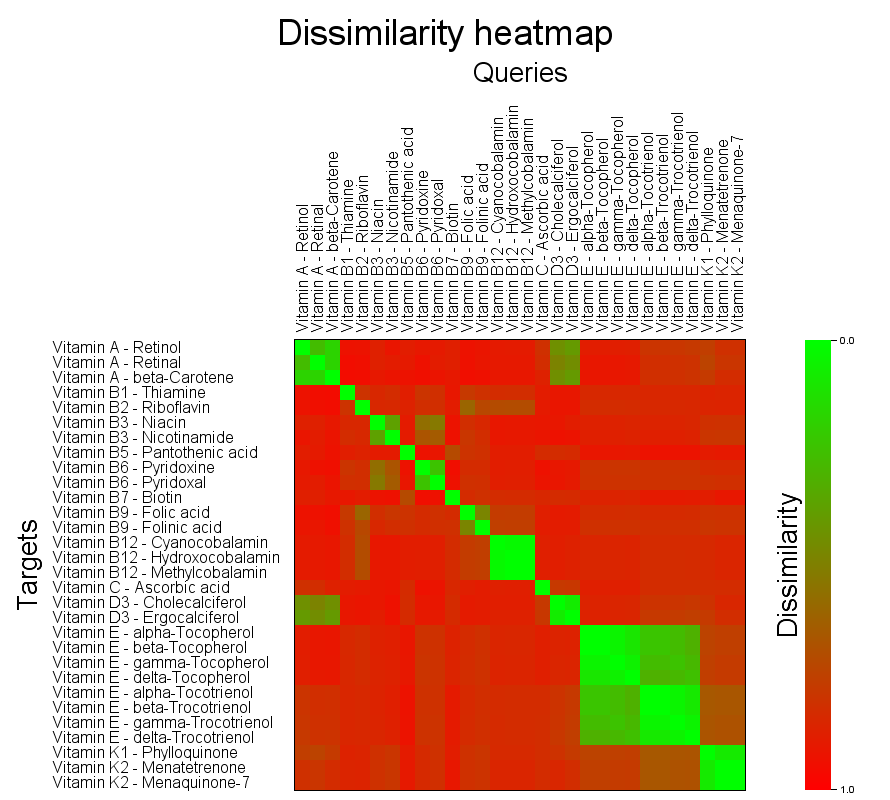
The generated image layout is adjusted to have larger than default cell sizes and enough space to accomodate the long structure ID strings of the dataset.
Breakdown of the arguments
| Command line part | Description |
|---|---|
-context createSimpleCfp7Context |
Descriptor (fingerprint) to be used. See Basic overview of the concepts of overlap analysis context for details. |
-qmf data/molecules/antibiotics/vitamins.smi |
Read queries from molecule file data/molecules/antibiotics/vitamins.smi, parse them and calculate descriptors according the context set. |
-qidname |
Use the molecule name field of the queries as IDs. |
-tmf data/molecules/antibiotics/vitamins.smi |
Read targets from molecule file data/molecules/antibiotics/vitamins.smi, parse them and calculate descriptors according the context set. |
-tidname |
Use the molecule name field of the targets as IDs. |
-mode FULLMATRIX |
Calculate and store the results of every query-target comparisons. |
-out vitamins-fullmatrix.txt |
Write textual results (dissimilarity matrix) to file vitamins-fullmatrix.txt. |
-heatmap-image vitamins-fullmatrix.png |
Create heatmap image from the search results and write it to file vitamins-fullmatrix.png. |
-heatmap-image-cellsize 15 |
Size the cells of the heatmap to 15 pixel * 15 pixel. This is an optional parameter. |
-heatmap-image-query-ids-length 250 |
Allow 250 pixels to print query IDs (the vitamins dataset contains relatively long molecule names). This is an optional parameter. |
-heatmap-image-query-ids-length 250 |
Allow 250 pixels to print target IDs. This is an optional parameter. |
Overlap of the antibiotics dataset with the essential medicines datatset
bin/searchStorage.sh \
-qmf data/molecules/antibiotics/antibiotics.smi \
-qidname \
-tmf data/molecules/who-essential-medicines/who-essential-medicines.smi \
-tidname \
-context createSimpleCfp7Context \
-mode MOSTSIMILARS \
-count 100 \
-maxdissim 0.15 \
-out antibiotics-vs-essentials-mostsimilars.txt \
-heatmap-image antibiotics-vs-essentials-mostsimilars.png \
-heatmap-image-cellsize 10 \
-heatmap-image-title-text "Antibiotics in the WHO Model List of Essential Medicines dataset" \
-heatmap-image-query-ids-length 100 \
-heatmap-image-query-label-text "Queries: List of antibiotics dataset" \
-heatmap-image-target-ids-length 200 \
-heatmap-image-target-label-text "Targets: WHO Model List of Essential Medicines dataset"
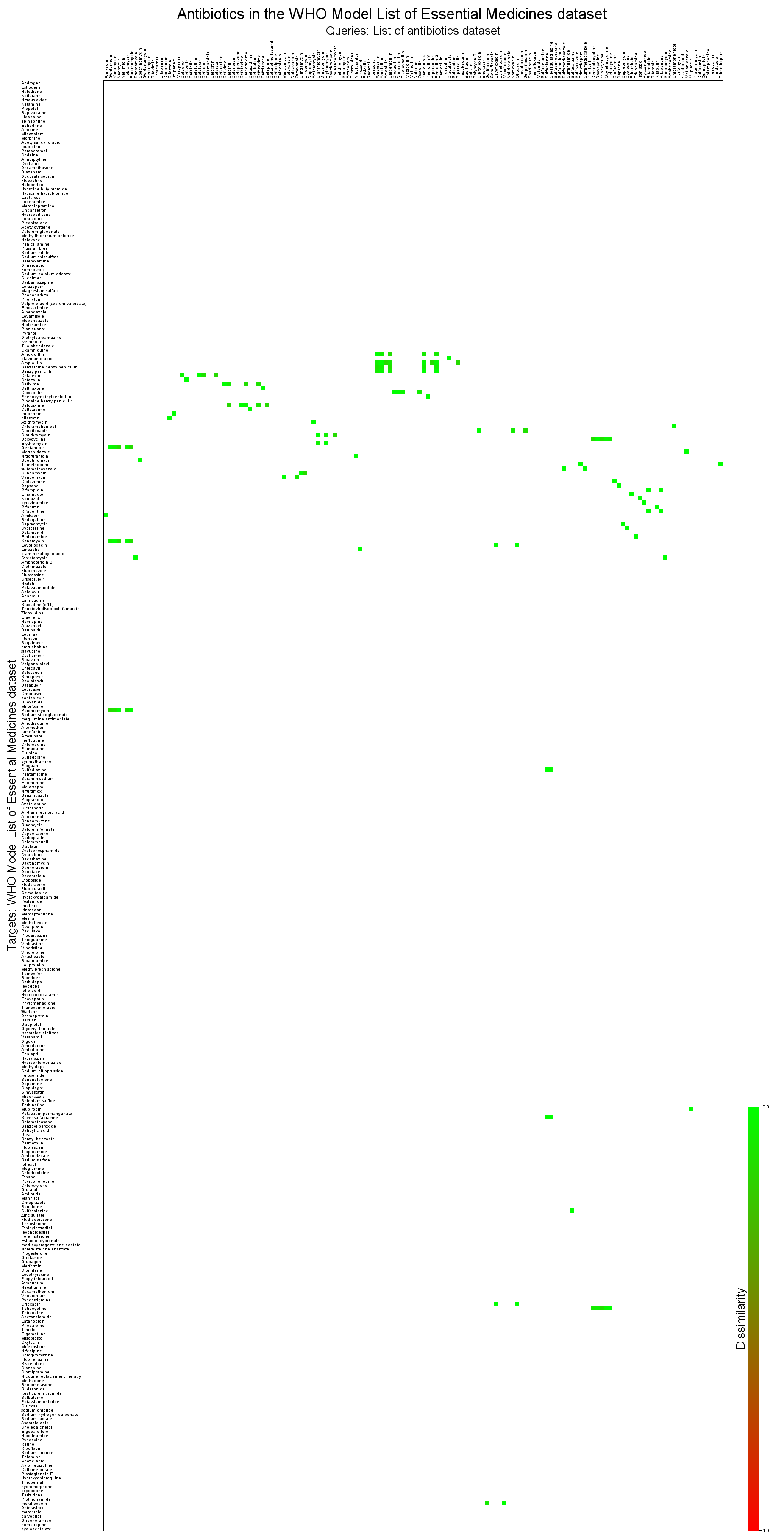
Breakdown of the arguments not used in the previous example
| Command line part | Description |
|---|---|
-mode MOSTSIMILARS |
Most similars search mode. At most the n most similar targets for each query are retrieved. |
-count 100 |
Set the number of maximal hits for each query to 100. |
-maxdissim 0.15 |
Targets with greater dissimilarity (smaller similarity) are rejected. |
-heatmap-image-title-text "...." |
Set the chart title. |
-heatmap-image-query-label-text "..." |
Set query labels. |
-heatmap-image-target-label-text "..." |
Set target labels. |
Self overlap of the drugbank-all dataset
gzip -dc data/molecules/drugbank/drugbank-common_name.smi.gz > drugbank.smi
bin/searchStorage.sh \
-qmf drugbank.smi \
-tmf drugbank.smi \
-context createSimpleCfp7Context \
-mode FULLMATRIX \
-out "" \
-heatmap-image drugbank-fullmatrix.png \
-heatmap-image-cellsize 1 \
-heatmap-image-title-text "Self overlap of the Drugbank-all dataset" \
-heatmap-image-query-label-text "" \
-heatmap-image-target-label-text ""
This dataset contains ~7k molecules, so the resulting image size is using 1 pixel by 1 pixel cells is larger than 50 Megapixels, 50 Megabytes in size. Resulting text output is not written (option -out "" used). Execution time of the command is expected to be around a minute. The output image is not available in this documentation.