Basic similarity search workflow
This is an example of using the supplied command line tools to generate descriptors for molecule sets and invoke similarity searches on them using molecule queries. Parts of the steps described below are implemented in script search-workflow.sh found in examples/ directory. This basic workflow consists of the following steps:
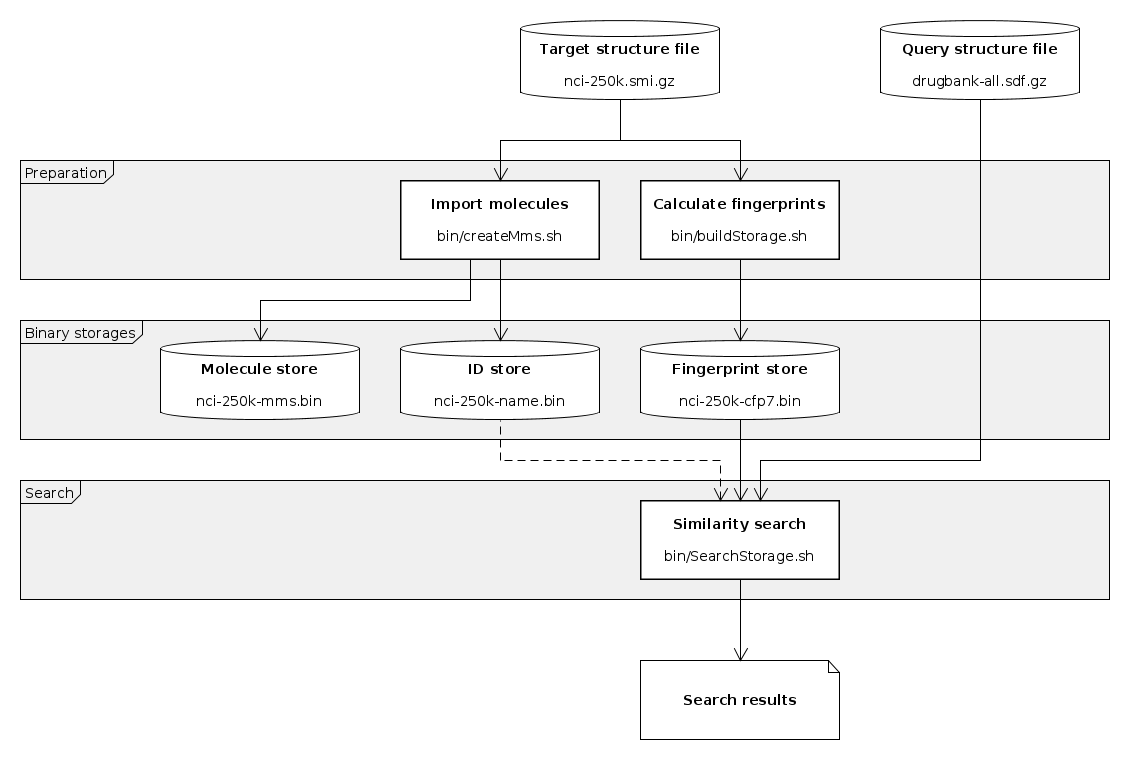
- Import molecules and IDs from structure file (creating master molecule storage)
- Calculate molecular descriptors to be used as targets for the search
- Invoke similarity search on prepared storages
- Diagnostic dump of prepared storages (optional)
For more details on the command line scripts involved see their description. For more details on performance see document Performance. An example for skipping the preparation steps and do on the fly descriptor calculation is also given. See also document Details on searchStorage.
Create master molecule storage
Master molecule storage used by other scripts (search) to retrieve structure sources and IDs. Structure IDs also stored in a similar data structure.
Notes
- These examples also create a serialized file containing molecule names (as ID) by specifying parameter
-name. Creating this file is optional. - Current version keep all parsed structure and the optional IDs/names in memory during processing.
- Since version 0.3.0 gzipped input files are automatically recognized. In some of the examples we use
gzipto decompress the the input file and read it from the standard input using command line option-in. When the input is read by the command line scripts (bin/<SCRIPT>.sh) the input file can be used directly.
Commands
# Retrieve IDs from SDF properties
gzip -dc data/molecules/drugbank/drugbank-all.sdf.gz | bin/createMms.sh \
-in - \
-out drugbank-all-mms.bin \
-prop COMMON_NAME:drugbank-all-commonname.bin
# Retrieve IDs from molecule name
gzip -dc data/molecules/nci/nci-250k.smi.gz | bin/createMms.sh \
-in - \
-out nci-250k-mms.bin \
-name nci-250k-name.bin
Breakdown of the invocations
| Command line part | Description |
|---|---|
gzip -dc <GZFILE> |
Decompress the content of gzip encoded file <GZFILE> and print it to the standard output. |
| |
Pipe the standard output of the previous command into the standard input of the following command. See http://www.tldp.org/LDP/abs/html/io-redirection.html for details. |
bin/createMms.sh |
Tool shipped to process input file and store structures and optionally IDs in a proprietary binary file readable by other tools. |
\ |
Sign that command is continued in the following line. |
-in <INPUT> |
Specify the location of input structures to process. |
- |
Specify standard input. |
-out <BINFILE> |
Specify the binary file for the master molecule storage to write. |
-prop <PROPNAME>:<BINFILE> |
Specify an SDF property <PROPNAME> to be extracted and stored in a binary file <BINFILE>. |
-name <BINFILE> |
Extract and store molecule name in a binary file <BINFILE>. |
Expected performance
Preprocessing the nci-250k dataset on recent desktop machine is expected to be done under a minute.
Calculate fingerprints
Generated descriptors (fingerprints) are stored in a binary file. This file will be read by search tool.
Notes
-
The settings regarding descriptor generation is determined by the specified
OverlapAnalysisContext. Examples below uses predefined contexts. Customization is possible using scripting hooks; see Basic overview of the concepts of overlap analysis context for details. -
Currently molecular descriptors are calculated from the original structure file. Current version does not recognize gzipped input files. We use
gzipto decompress the the input file and read it from the standard input using command line option-in.
Commands
gzip -dc data/molecules/drugbank/drugbank-all.sdf.gz | bin/buildStorage.sh \
-context createSimpleCfp7Context \
-in - \
-out drugbank-all-cfp7.bin
gzip -dc data/molecules/nci/nci-250k.smi.gz | bin/buildStorage.sh \
-context createSimpleCfp7Context \
-in - \
-out nci-250k-cfp7.bin
Expected performance
Fingerprint calculation for the nci-250k dataset on a recent desktop machine is expected to be done well under a minute.
Breakdown of the invocations
| Command line part | Description |
|---|---|
bin/buildStorage.sh |
Tool to process structure file input, calculate molecular descriptors (fingerprints) and store them in a binary file. |
-context <CONTEXT> |
Specify molecular descriptor, default comparison metric and other parameters to be used during calculation and later search. For details see document Basic overview of the concepts of overlap analysis context. |
-in <INFILE> |
Structure file to process. |
-out <BINFILE> |
Binary file containg calculated descriptors to write. |
Invoke similarity search from command line
By default targets are identified by their master index (0-based index in the input structure file). If serialized id or name storage (created with master molecule storage) is specified then the stored ID (or name) is retrieved and printed.
Commands
# Launch a simple search against the NCI database with a query molecule specified as a SMILES string
bin/searchStorage.sh \
-frombytes nci-250k-cfp7.bin \
-qm "C1CCCC1"
# Use previously extracted IDs
bin/searchStorage.sh \
-frombytes nci-250k-cfp7.bin \
-idstorage nci-250k-name.bin \
-qm "C1CCCC1"
# Find the 10 most similar structures
bin/searchStorage.sh \
-frombytes nci-250k-cfp7.bin \
-idstorage nci-250k-name.bin \
-mode MOSTSIMILARS \
-count 10 \
-qm "C1CCCC1"
# Search most similar from the NCI database for each of the first 10 of the Drugbank dataset
gzip -dc data/molecules/drugbank/drugbank-all.sdf.gz | head -10 | bin/searchStorage.sh \
-frombytes nci-250k-cfp7.bin \
-idstorage nci-250k-name.bin \
-qf -
Breakdown of the invocations
| Command line part | Description |
|---|---|
bin/searchStorage.sh |
Tool to invoke similarity searches against molecular descriptors stored in a binary file previously generated by buildStorage. |
-frombytes <BINFILE> |
Binary file containing molecular descriptors. |
-idstorage <BINFILE> |
Read target IDs from specified location. |
-qm <QMOLSOURCE> |
Import molecule from source <QMOLSOURCE> and use it as a query. |
-qf <QUERY> |
Import query molecules from specified location. |
-qf - |
Import query molecules from standard input. |
-mode MOSTSIMILARS |
Find the n most similar molecules for each query. |
-count 10 |
Specify the max number of most similar structures to find. |
Expected performance
Execution time of the above runs is expected to be in the few seconds range.
Diagnostics: dump contents of the serialized storages
Tool dumpStorage reads spcified binary files and prints an overview of their contents. Note that the given storage is fully read into memory (regardless of the printed line count).
Command
bin/dumpStorage.sh -in drugbank-all-cfp7.bin -in drugbank-all-mms.bin -in drugbank-all-commonname.bin
On the fly descriptor calculations
The example above use tools createMms and buildStorage to prepare descriptors and IDs for later search or for exposing through Web UI / REST API. It is possible to skip this preparation steps and let searchStorage to do the calculation. For further information on parametrization of searchStorage see Details on searchStorage.
# Find the 10 most similar structures using asymmetric tversky with on the fly descriptor calculation
tabs 40
gzip -dc gzip -dc data/molecules/drugbank/drugbank-all.sdf.gz | bin/searchStorage.sh \
-context createSimpleCfp7Context \
-metric "tversky,coeffT:0.01,coeffQ:0.99" \
-tmf - \
-tidprop COMMON_NAME \
-mode MOSTSIMILARS \
-count 10 \
-qm "O1CC1 epoxy" \
-qidname
The output:
Query Target Dissimilarity
epoxy Sevelamer 0.019607843137254943
epoxy Colestipol 0.027946537059538312
epoxy Fosfomycin 0.03264812575574361
epoxy 3-Oxiran-2ylalanine 0.03730445246690739
epoxy R-Styrene Oxide 0.03961584633853543
epoxy Oxiranpseudoglucose 0.04534606205250602
epoxy D-Limonene 1,2-Epoxide 0.04534606205250602
epoxy 3,4-Epoxybutyl-Alpha-D-Glucopyranoside 0.06432748538011701
epoxy (R)-4-Nitrostyrene oxide 0.06868451688009314
epoxy (S)-4-Nitrostyrene oxide 0.06868451688009314
Breakdown of the invocations
| Command line part | Description |
|---|---|
tabs 40 |
Set tab stops of the terminal to 40 characters. This ensures that the columns of the ouptut are visually aligned. See https://linux.die.net/man/1/tabs. |
gzip -dc <GZFILE> |
Decompress the content of gzip encoded file <GZFILE> and print it to the standard output. |
| |
Pipe the standard output of the previous command into the standard input of the following command. See http://www.tldp.org/LDP/abs/html/io-redirection.html for details. |
bin/searchStorage.sh |
Tool to invoke similarity searches against molecular descriptors stored in a binary file previously generated by buildStorage or generated on the fly. |
\ |
Sign that command is continued in the following line. |
-context <CONTEXT> |
Specify molecular descriptor, default comparison metric and other parameters to be used during calculation and later search. For details see document Basic overview of the concepts of overlap analysis context. |
-metric <METRIC> |
Customize comparison metric. |
tversky,coeffT:0.01,coeffQ:0.99 |
Asymmetric tversky metric with parameters where query only features are highly penalized, while target only features are slightly penalized. |
-tmf <MOLFILE> |
Read and parse targets from a molecule file |
-tmf - |
Use stdin to read the target molecules from |
-tidprop <PROPNAME> |
Extract target IDs from the given property of the parsed target molecules |
-tidprop COMMON_NAME |
Property name to use for target IDs |
-mode MOSTSIMILARS |
Find the n most similar molecules for each query. |
-count 10 |
Specify the max number of most similar structures to find. |
-qm <QMOLSOURCE> |
Import molecule from source <QMOLSOURCE> and use it as a query. |
-qm "O1CC1 epoxy" |
SMILES structure source with molecule name specified. |
-qidname |
Use molecule name of query molecule(s) as query IDs . |
Output formatting
By default the dissimilarity values uses Java Double formatting. Using option -out-format <FORMAT> a custom formatting can be specified which delegates to Java java.text.Format. The following example use %.3f for a fixed 3 digit precision:
# Find the 5 most similar structures using asymmetric tversky with on the fly descriptor calculation
tabs 25
gzip -dc gzip -dc data/molecules/drugbank/drugbank-all.sdf.gz | bin/searchStorage.sh \
-context createSimpleCfp7Context \
-metric "tversky,coeffT:0.01,coeffQ:0.99" \
-tmf - \
-tidprop COMMON_NAME \
-mode MOSTSIMILARS \
-count 5 \
-qm "O1CC1 epoxy" \
-qidname \
-out-numeric-format "%.3f"
The output:
Query Target Dissimilarity
epoxy Sevelamer 0.020
epoxy Colestipol 0.028
epoxy Fosfomycin 0.033
epoxy 3-Oxiran-2ylalanine 0.037
epoxy R-Styrene Oxide 0.040
Heatmap visualization
As an experimental feature a heatmap of the search results can be calculated using options -heatmap-image <FILE> and -heatmap-image-cellsize <CELLSIZE>. Search modes MOSTSIMILAR, MOSTSIMILARS and FULLMATRIX are all supported. Please note that heatmap rendering is not recommended for very large datasets. The approximate pixel count of the resulting image is <QUERIES> * <TARGETS> * <CELLSIZE> * <CELLSIZE> which is recommended to be kept below a few tens of megapixels.
Self overlap of the vitamins dataset
bin/searchStorage.sh \
-context createSimpleCfp7Context \
-qmf data/molecules/vitamins/vitamins.smi \
-qidname \
-tmf data/molecules/vitamins/vitamins.smi \
-tidname \
-mode FULLMATRIX \
-out vitamins-fullmatrix.txt \
-heatmap-image vitamins-fullmatrix.png \
-heatmap-image-cellsize 15 \
-heatmap-image-query-ids-length 250 \
-heatmap-image-target-ids-length 250
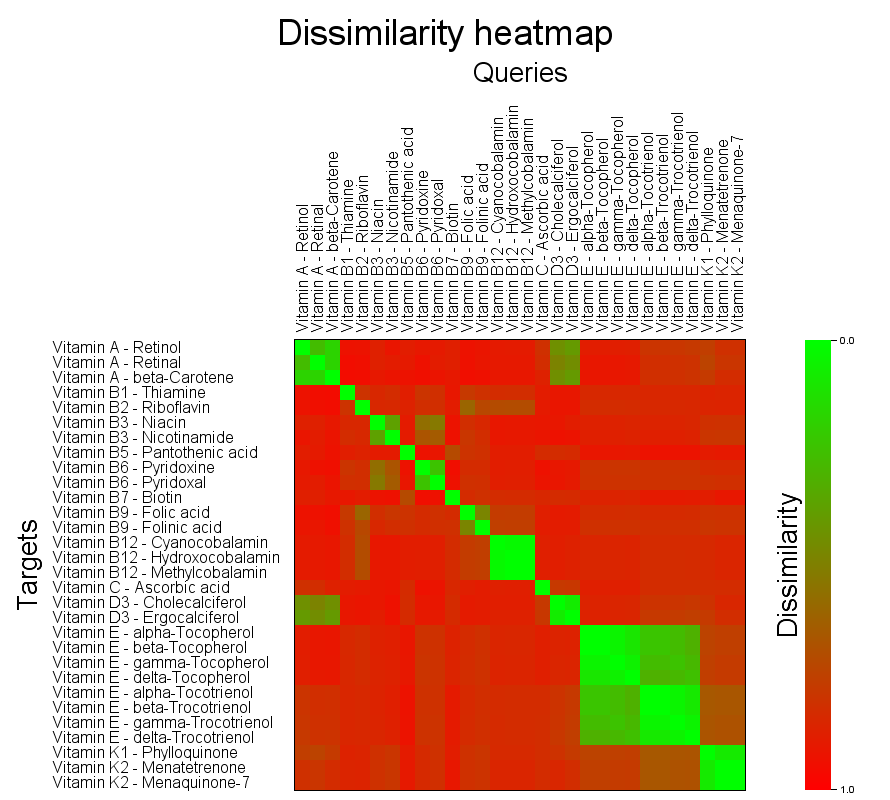
The generated image layout is adjusted to have larger than default cell sizes and enough space to accomodate the long structure ID strings of the dataset. For details on the heatmap image generation see document Details on searchStorage.